CLASP - Contrastive Language-Aminoacid Sequence Pretraining
Repository for creating models pretrained on language and aminoacid sequences similar to ConVIRT, CLIP, and ALIGN.
To dos
- Finish current big run with unprocessed data.
- Evaluate the model on zero-shot tasks. See the introduction inference notebook based on the checkpoints from below.
- Train new model with processed data. See the preprocessing scripts in the
preprocdirectory.
If you are interested in this project and you want to contribute feel free to get in touch (see Discussion below).
Updates
Run 44-48 with UniProt full dataset (~213 mio samples):

Model checkpoint:
230t steps train: loss: 1.067, acc: 0.702, valid-id: loss: 1.587: acc: 0.561, valid-ood: loss: 2.545: acc: 0.352 gdrive download (~1GB)
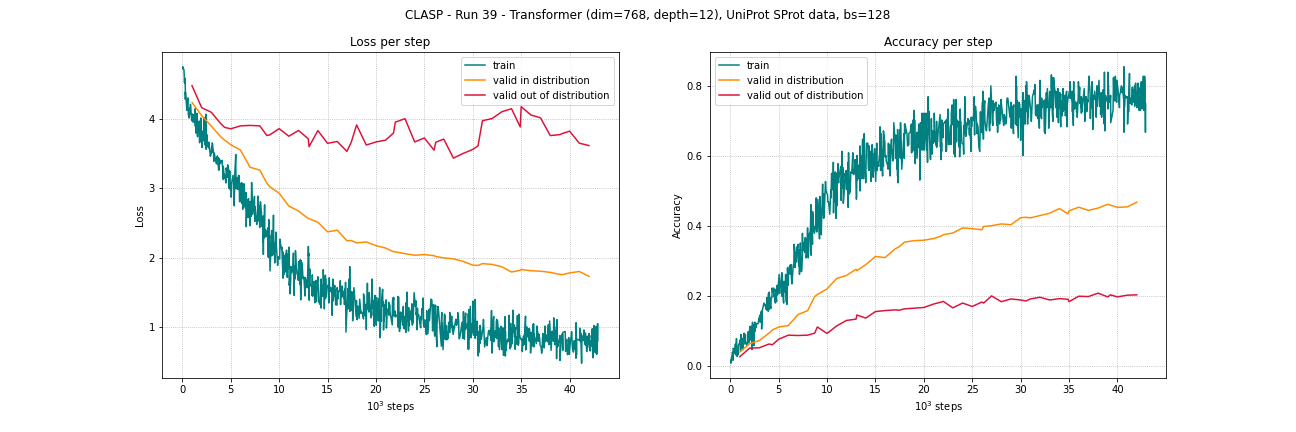
Run 39 with UniProt SProt dataset (~0.5 mio samples):
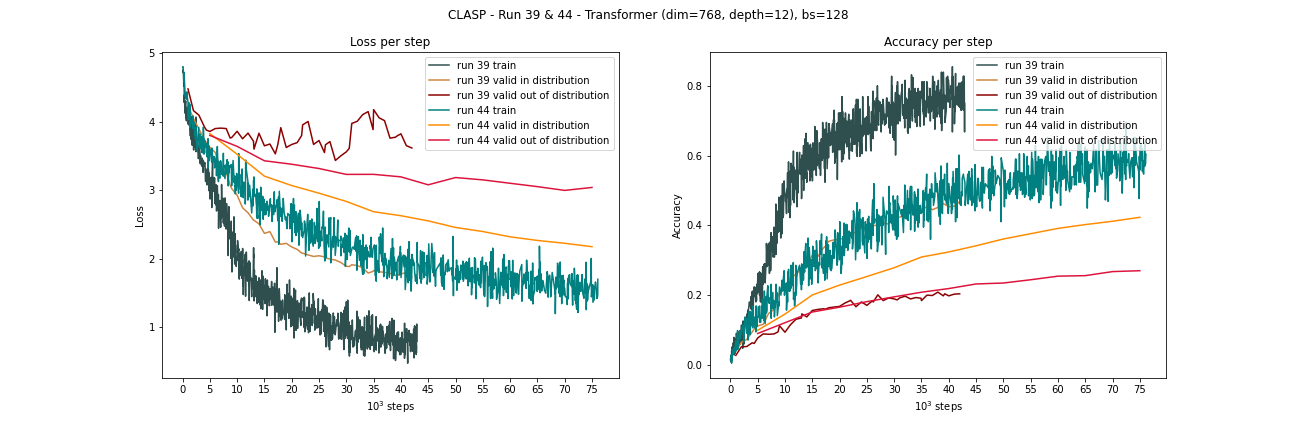
Comparison run 39 and run 44 from above:
Work in progress, more updates soon!
Discussion
EleutherAI discord server or alphafold2 discord server.
Ping @MicPie
Data
For model training the data provided by UniProt is used.
Requirements
You can install the requirements with the following
$ python setup.py install --user
Then, you must install Microsoft's sparse attention CUDA kernel with the following two steps.
$ sh install_deepspeed.sh
Next, you need to pip install the package triton
$ pip install triton
If both of the above succeeded, now you can train your long biosequences with CLASP
Usage
import torch
from torch.optim import Adam
from clasp import CLASP, Transformer, tokenize
# instantiate the attention models for text and bioseq
text_enc = Transformer(
num_tokens = 20000,
dim = 512,
depth = 6,
seq_len = 1024,
reversible = True
)
bioseq_enc = Transformer(
num_tokens = 21,
dim = 512,
depth = 6,
seq_len = 512,
sparse_attn = True,
reversible = True
)
# clasp (CLIP) trainer
clasp = CLASP(
text_encoder = text_enc,
bioseq_encoder = bioseq_enc
)
# data
text, text_mask = tokenize(['Spike protein S2: HAMAP-Rule:MF_04099'], context_length = 1024, return_mask = True)
bioseq = torch.randint(0, 21, (1, 511)) # when using sparse attention, should be 1 less than the sequence length
bioseq_mask = torch.ones_like(bioseq).bool()
# do the below with large batch sizes for many many iterations
opt = Adam(clasp.parameters(), lr = 3e-4)
loss = clasp(
text,
bioseq,
text_mask = text_mask,
bioseq_mask = bioseq_mask,
return_loss = True # set return loss to True
)
loss.backward()
Once trained
scores = clasp(
texts,
bio_seq,
text_mask = text_mask,
bioseq_mask = bioseq_mask
)
Resources
See interesting resources (feel free to add interesting material that could be useful).
Acknowledgements
This project is supported by EleutherAI.
Citations
@article{zhang2020contrastive,
title={Contrastive learning of medical visual representations from paired images and text},
author={Zhang, Yuhao and Jiang, Hang and Miura, Yasuhide and Manning, Christopher D and Langlotz, Curtis P},
journal={arXiv preprint arXiv:2010.00747},
year={2020}
}
OpenAI blog post "CLIP: Connecting Text and Images"
@article{radford2021learning,
title={Learning transferable visual models from natural language supervision},
author={Radford, Alec and Kim, Jong Wook and Hallacy, Chris and Ramesh, Aditya and Goh, Gabriel and Agarwal, Sandhini and Sastry, Girish and Askell, Amanda and Mishkin, Pamela and Clark, Jack and others},
journal={arXiv preprint arXiv:2103.00020},
year={2021}
}
@article{jia2021scaling,
title={Scaling Up Visual and Vision-Language Representation Learning With Noisy Text Supervision},
author={Jia, Chao and Yang, Yinfei and Xia, Ye and Chen, Yi-Ting and Parekh, Zarana and Pham, Hieu and Le, Quoc V and Sung, Yunhsuan and Li, Zhen and Duerig, Tom},
journal={arXiv preprint arXiv:2102.05918},
year={2021}
}










