DNA
This repository provides the code of our paper: Blockwisely Supervised Neural Architecture Search with Knowledge Distillation.
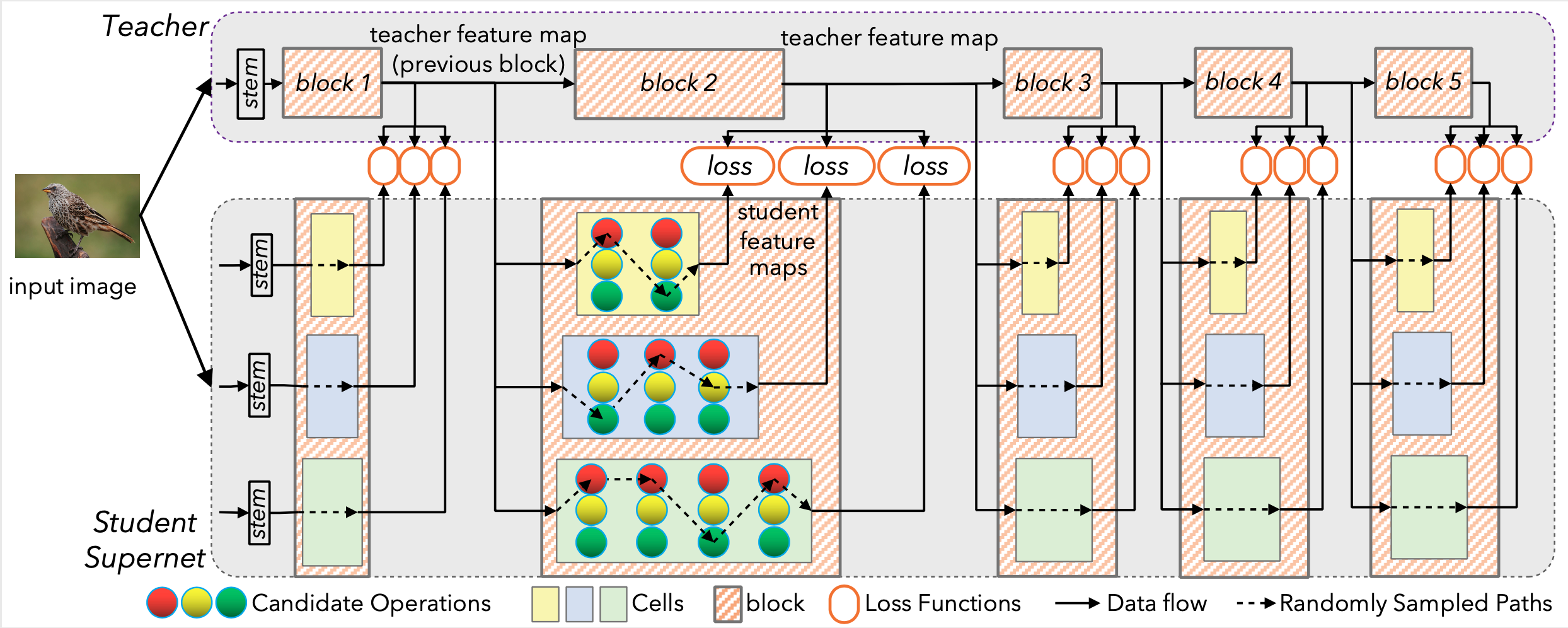
Illustration of DNA. Each cell of the supernet is trained independently to mimic the behavior of the corresponding teacher block.

Comparison of model ranking for DNA vs. DARTS, SPOS and MnasNet under two different hyper-parameters.
Our Trained Models
-
Our searched models have been trained from scratch and can be found in: https://drive.google.com/drive/folders/1Oqc2gq8YysrJq2i6RmPMLKqheGfB9fWH.
-
Here is a summary of our searched models:
Model FLOPs Params Acc@1 Acc@5 DNA-a 348M 4.2M 77.1% 93.3% DNA-b 394M 4.9M 77.5% 93.3% DNA-c 466M 5.3M 77.8% 93.7% DNA-d 611M 6.4M 78.4% 94.0%

Usage
1. Requirements
- Install PyTorch (pytorch.org)
- Install third-party requirements
pip install timm==0.1.14We use this pytorch-image-models codebase to validate our models.
- Download the ImageNet dataset and move validation images to labeled subfolders
- To do this, you can use the following script: https://raw.githubusercontent.com/soumith/imagenetloader.torch/master/valprep.shvalprep.sh
- Only the validation set is needed in the evaluation process.
2. Searching
The code for supernet training, evaluation and searching is under searching directory.
cd searching
i) Train & evaluate the block-wise supernet with knowledge distillation
- Modify datadir in
initialize/data.yamlto your ImageNet path. - Modify nproc_per_node in
dist_train.shto suit your GPU number. The default batch size is 64 for 8 GPUs, you can change batch size and learning rate ininitialize/train_pipeline.yaml - By default, the supernet will be trained sequentially from stage 1 to stage 6 and evaluate after each stage. This will take about 2 days on 8 GPUs with EfficientNet B7 being the teacher. Resuming from checkpoints is supported. You can also change
start_stageininitialize/train_pipeline.yamlto force start from a intermediate stage without loading checkpoint. sh dist_train.sh
ii) Search for the best architecture under constraint.
Our traversal search can handle a search space with 6 ops in each layer, 6 layers in each stage, 6 stages in total. A search process like this should finish in half an hour with a single cpu. To perform search over a larger search space, you can manually divide the search space or use other search algorithms such as Evolution Algorithms to process our evaluated architecture potential files.
- Copy the path of architecture potential files generated in step i) to
potential_yamlinprocess_potential.py. Modify the constraint inprocess_potential.py. python process_potential.py
3. Retraining
The retraining code is simplified from the repo: pytorch-image-models and is under retraining directory.
-
cd retraining -
Retrain our models or your searched models
- Modify the
run_example.sh: change data path and hyper-params according to your requirements - Add your searched model architecture to
model.py. You can also use our searched and predefined DNA models. sh run_example.sh
- Modify the
-
You can evaluate our models with the following command:
python validate.py PATH/TO/ImageNet/validation --model DNA_a --checkpoint PATH/TO/model.pth.tarPATH/TO/ImageNet/validationshould be replaced by your validation data path.--model:DNA_acan be replaced byDNA_b,DNA_c,DNA_dfor our different models.--checkpoint: Suggest the path of your downloaded checkpoint here.



