CaraNet
CaraNet: Context Axial Reverse Attention Network for Small Medical Objects Segmentation
This repository contains the implementation of a novel attention based network (CaraNet) to segment the polyp (CVC-T, CVC-ClinicDB, CVC-ColonDB, ETIS and Kvasir) and brain tumor (BraTS). The CaraNet show great overall segmentation performance (mean dice) on polyp and brain tumor, but also show great performance on small medical objects (small polyps and brain tumors) segmentation.
The technique report is here: CaraNet
Architecture of CaraNet
Backbone
We use Res2Net as our backbone.
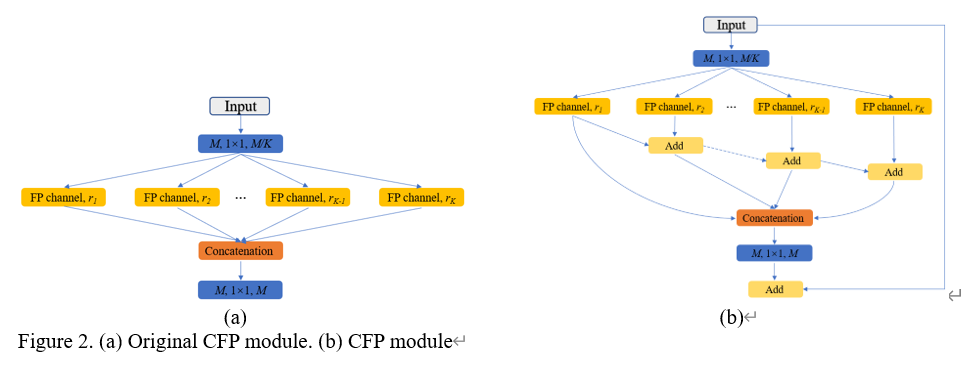
Context module
We choose our CFP module as context module, and choose the dilation rate is 8. For the details of CFP module you can find here: CFPNet. The architecture of CFP module as shown in following figure:
Axial Reverse Attention
As shown in architecture of CaraNet, the Axial Reverse Attention (A-RA) module contains two routes: 1) Reverse attention; 2) Axial-attention.
Installation & Usage
Enviroment
- Enviroment: Python 3.6;
- Install some packages:
conda install pytorch==1.1.0 torchvision==0.3.0 cudatoolkit=10.0 -c pytorch
conda install opencv-python pillow numpy matplotlib
- Clone this repository
git clone https://github.com/AngeLouCN/CaraNet
Training
- Download the training and texting dataset from this link: Experiment Dataset
- Change the --train_path & --test_path in Train.py
- Run
Train.py - Testing dataset is ordered as follow:
|-- TestDataset
| |-- CVC-300
| | |-- images
| | |-- masks
| |-- CVC-ClinicDB
| | |-- images
| | |-- masks
| |-- CVC-ColonDB
| | |-- images
| | |-- masks
| |-- ETIS-LaribPolypDB
| | |-- images
| | |-- masks
| |-- Kvasir
| |-- images
| |-- masks
Testing
- Change the data_path in Test.py
Evaluation
- Change the image_root and gt_root in eval_Kvasir.py
- You can also run the matlab code in eval fold, it contains other four measurement metrics results.
- You can download the segmentation maps of CaraNet from this link: CaraNet
dice_average.mis to compute the averaged dice values according to sizes of objects, for small area analysis.
Segmentation Results
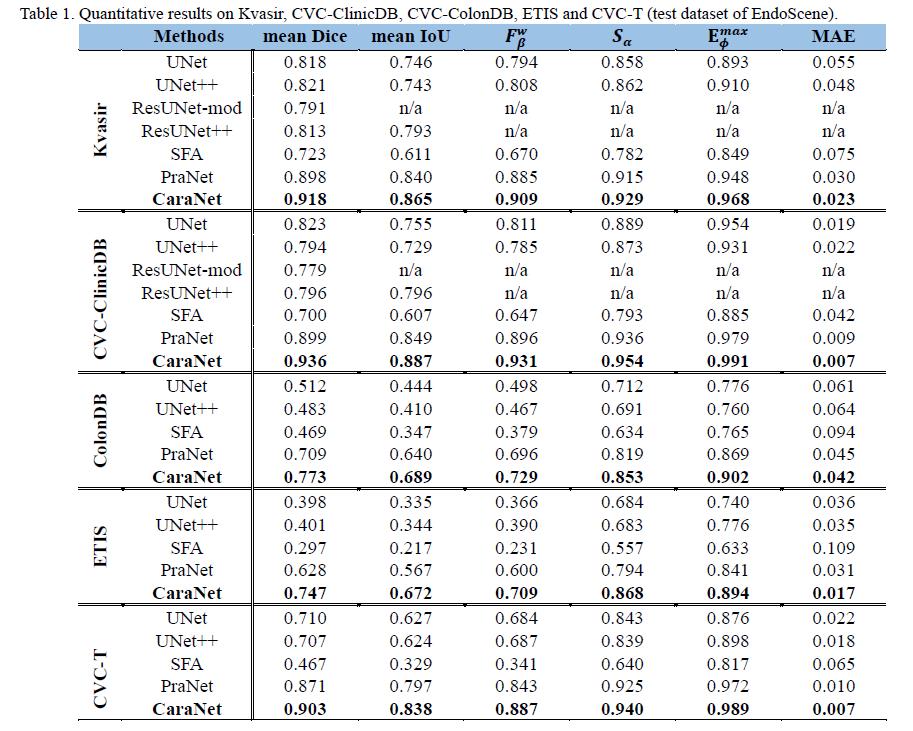
- Polyp Segmentation Results
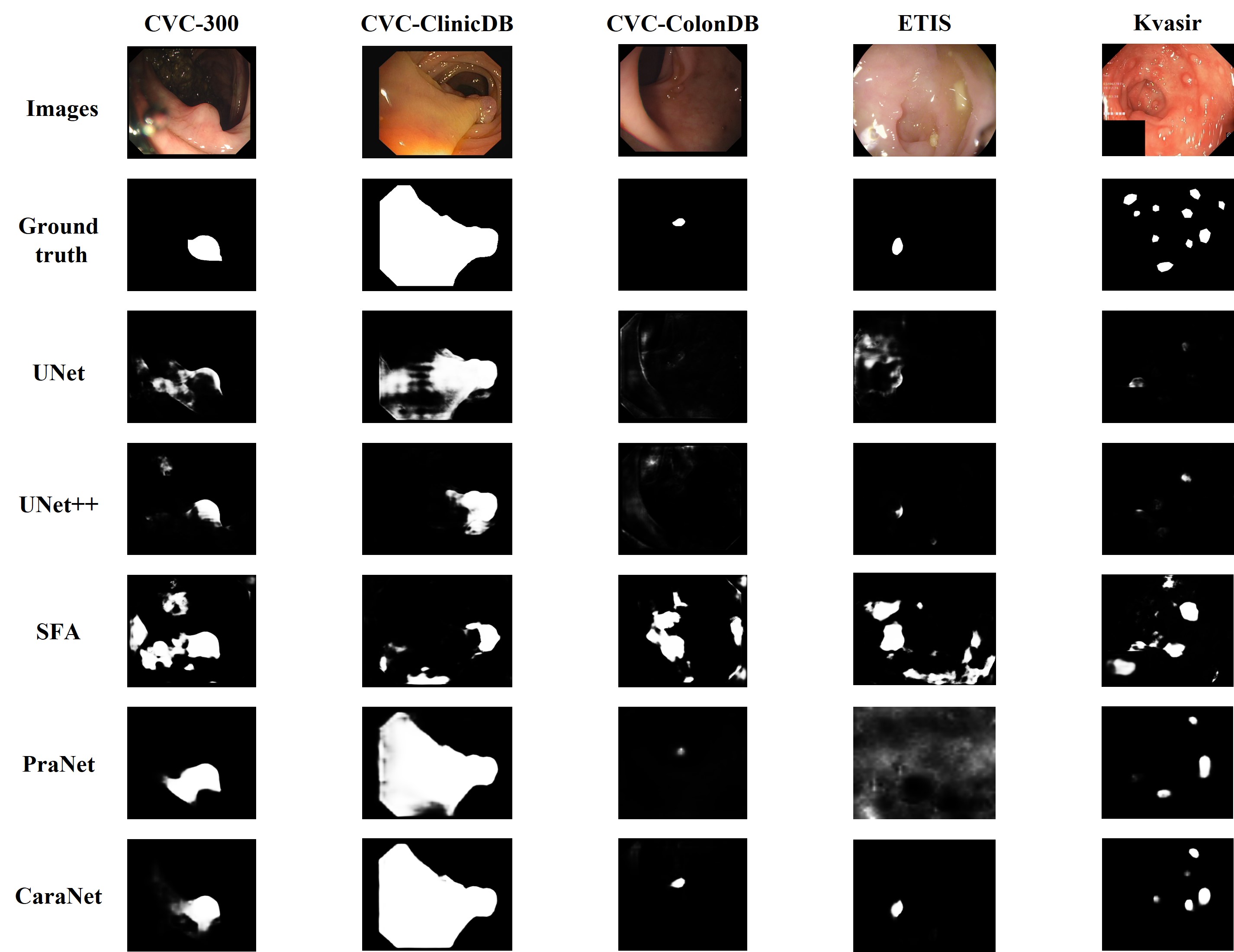
- Conditions of test datasets:
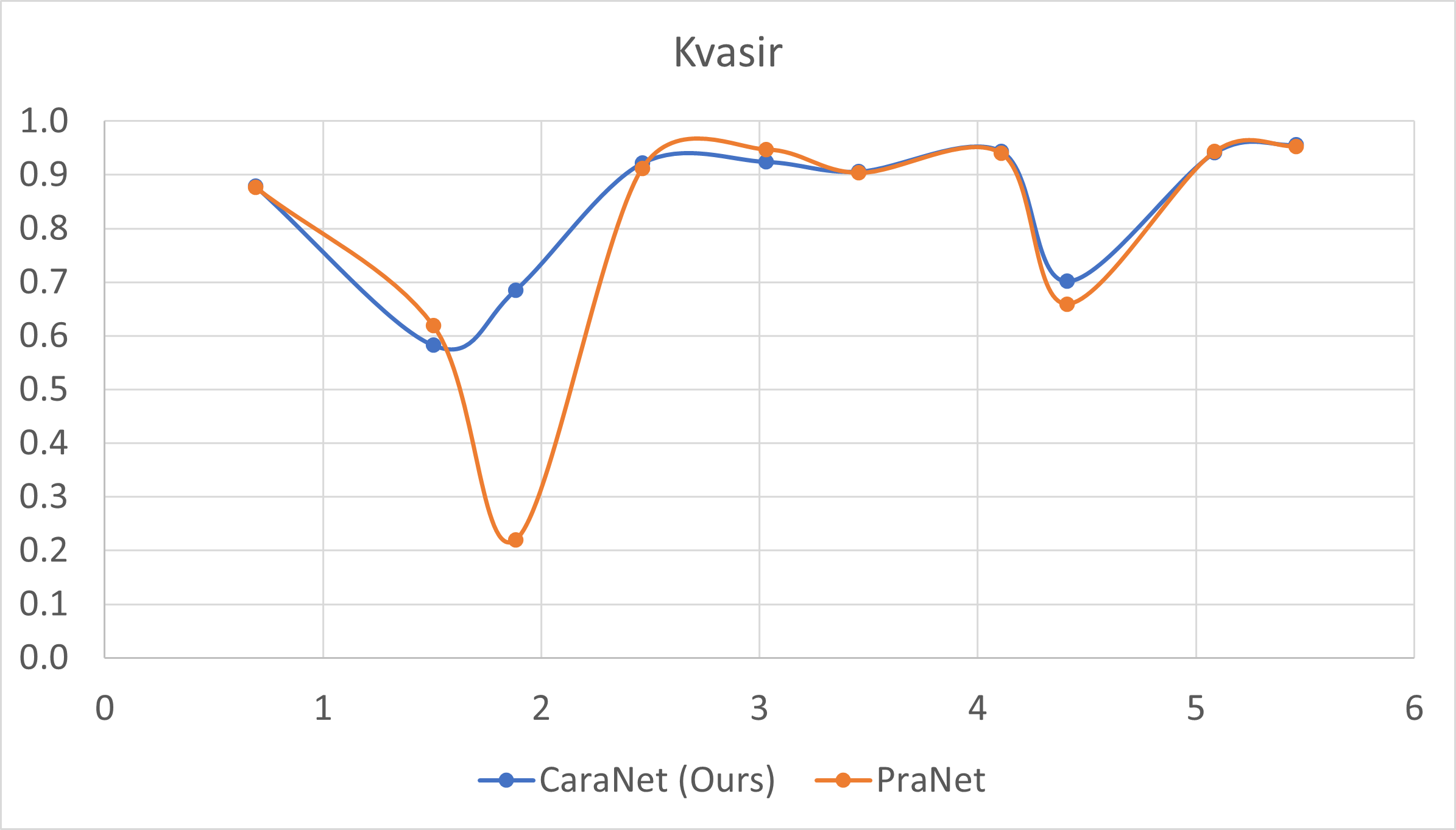
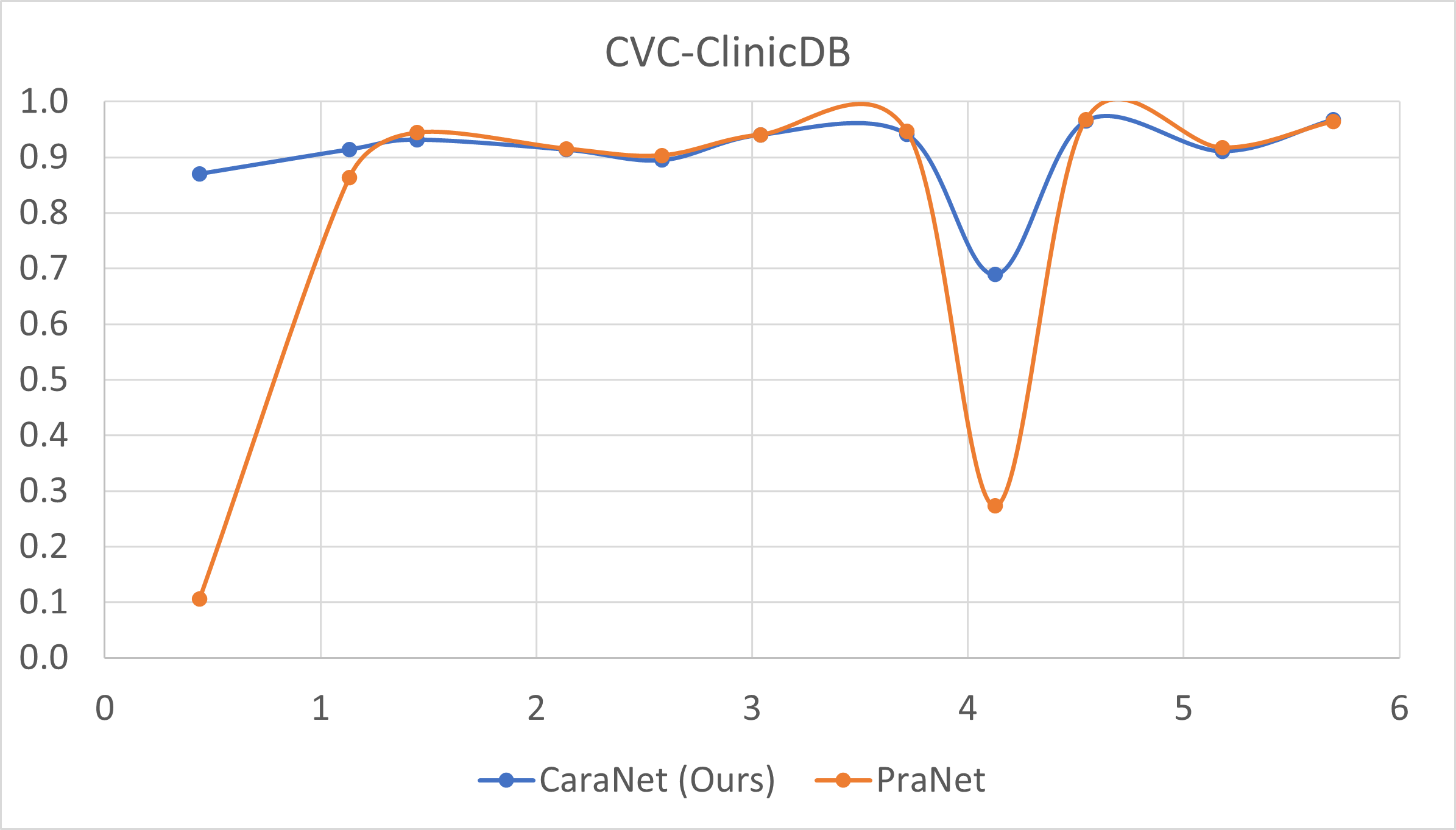
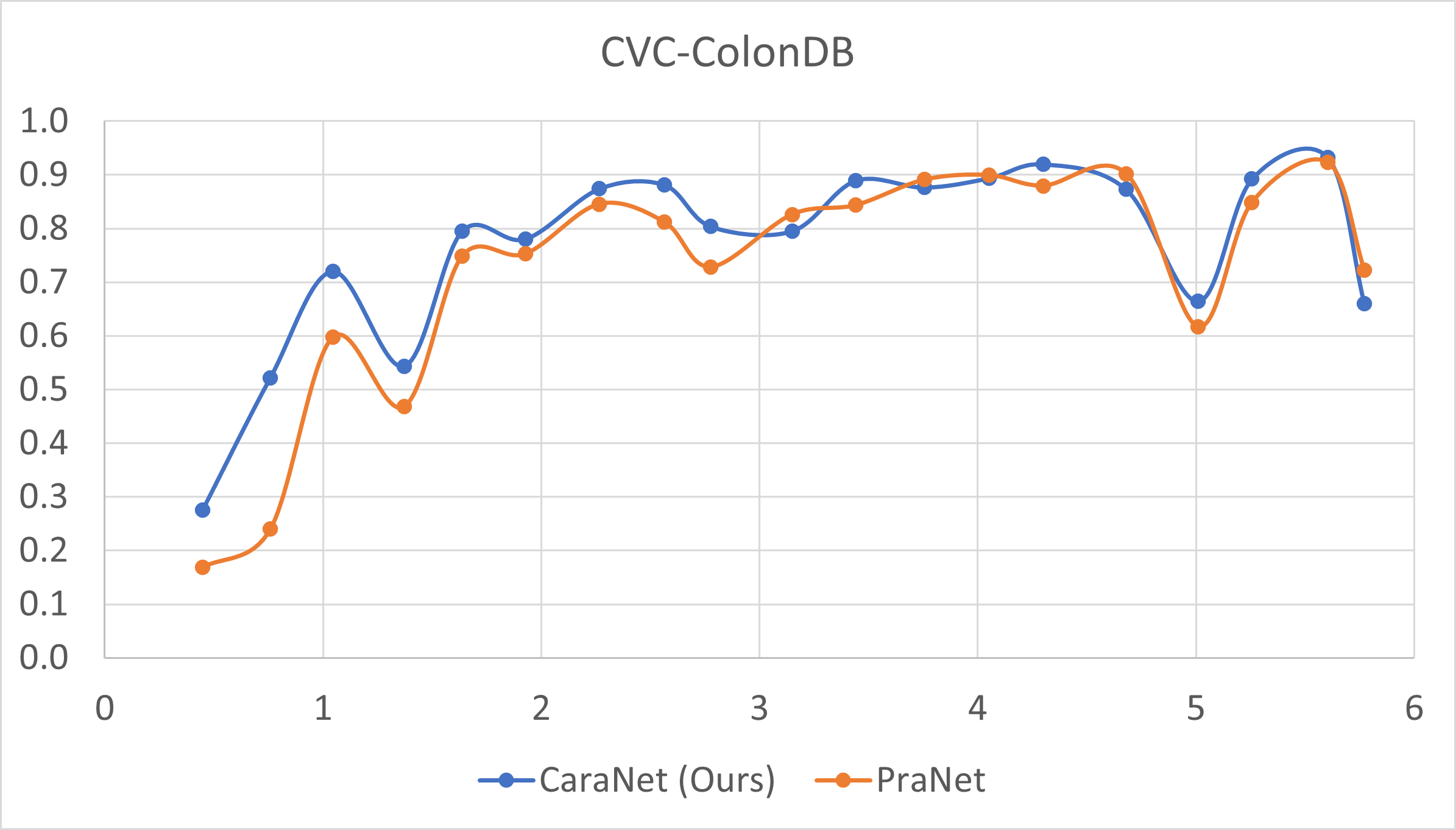
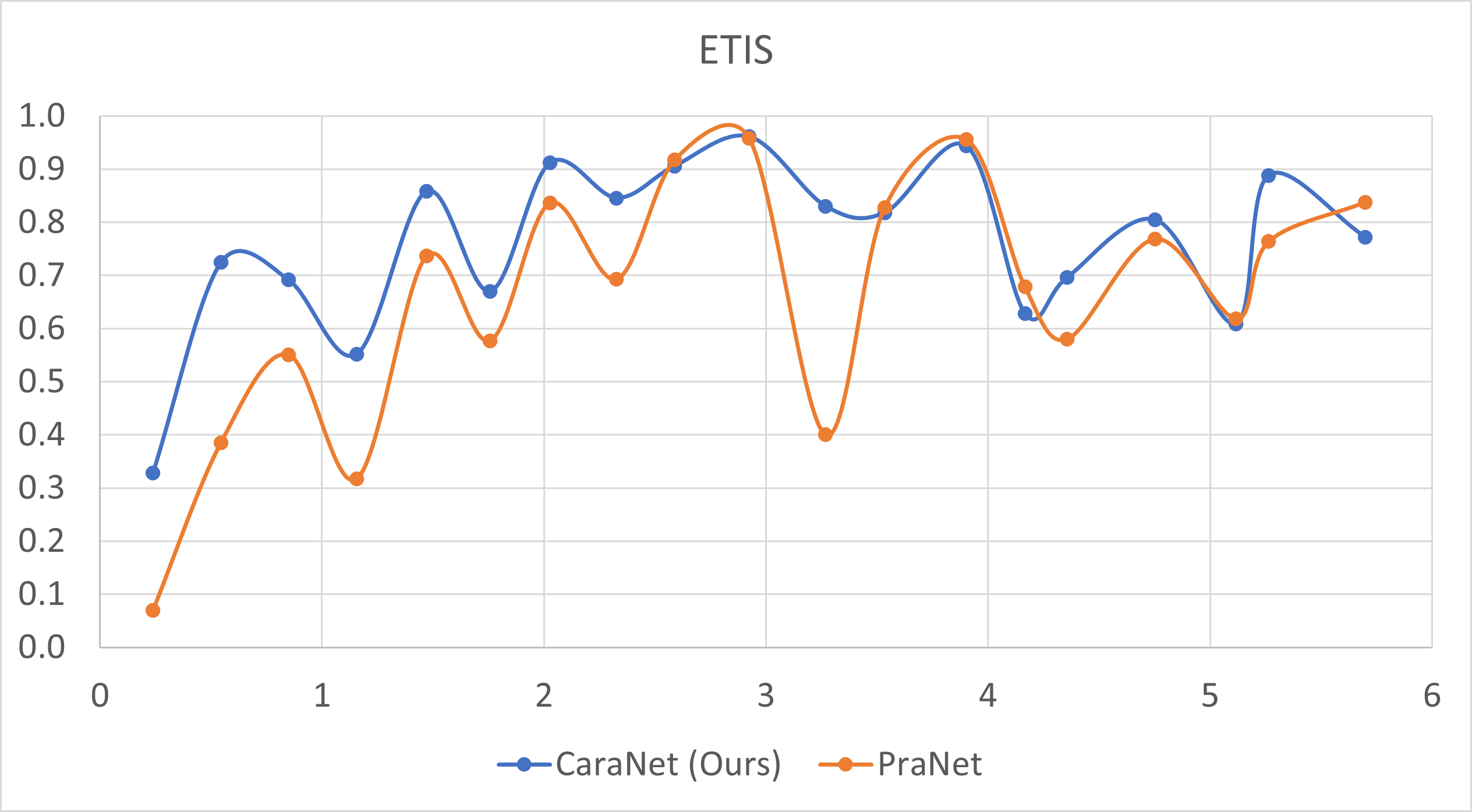
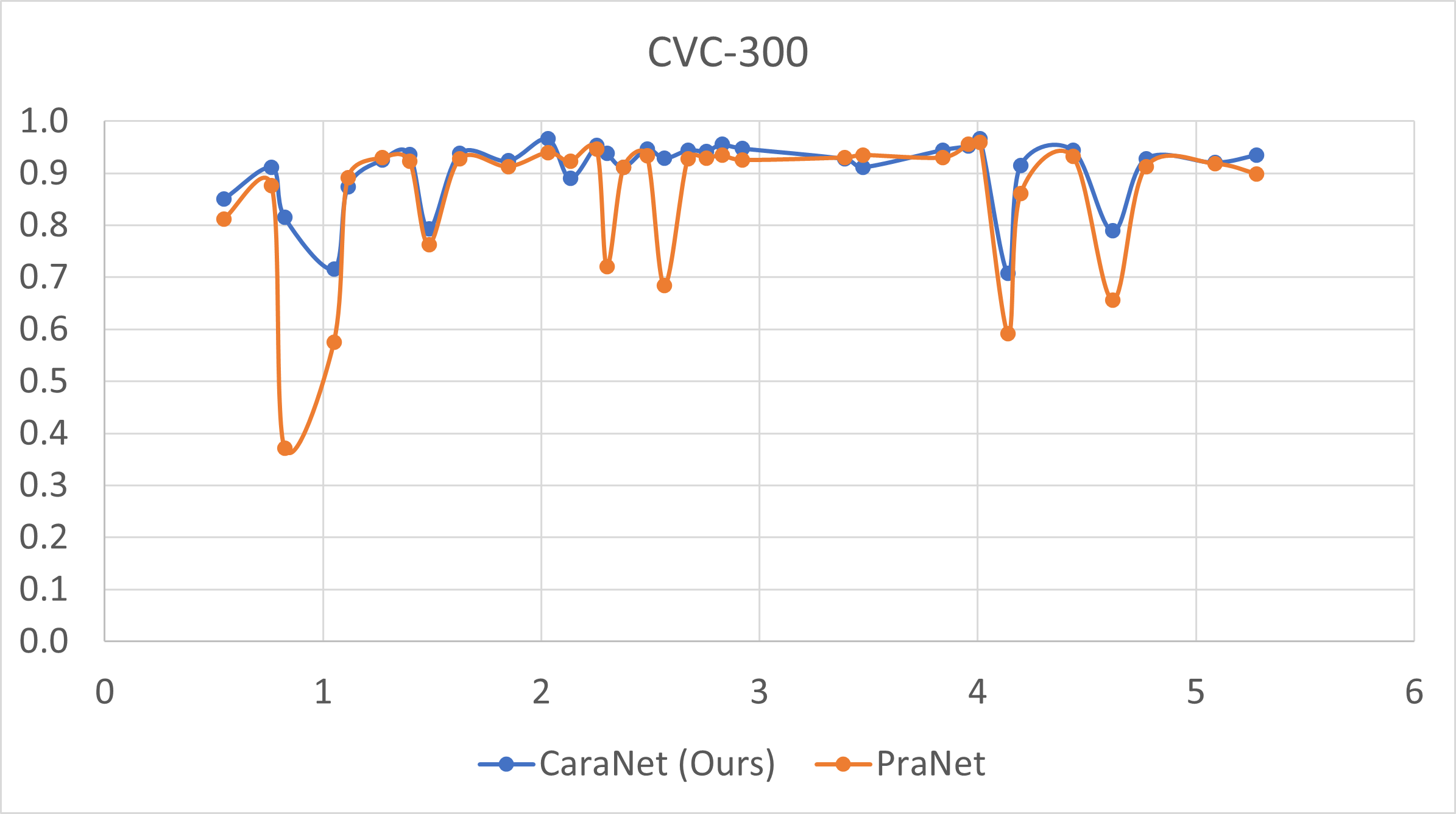
- Small polyp analysis
The x-axis is the proportion size (%) of polyp; y-axis is the average mean dice coefficient.
Brain Tumor Segmentation
- Dataset
| BraTS input | Segmentation truth |
|---|---|
 |
 |
- Results
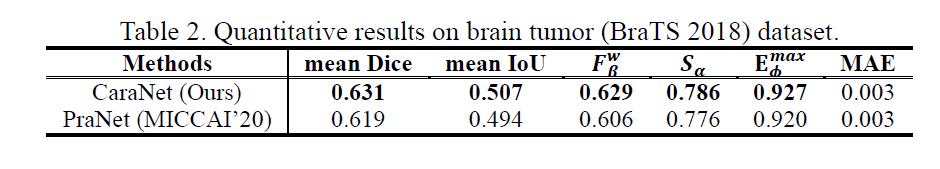
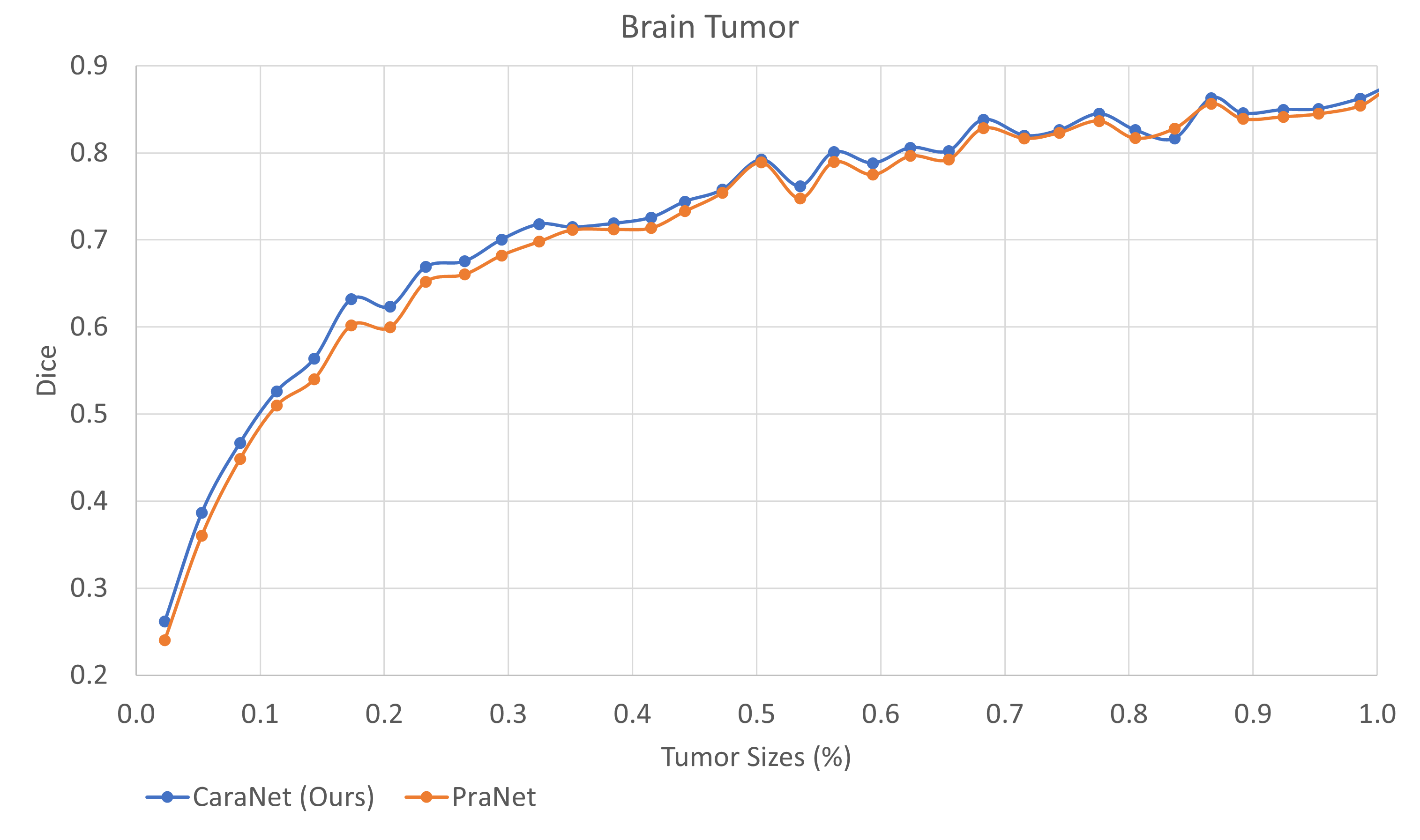
- Small tumor analysis
For very small areas (<1%):
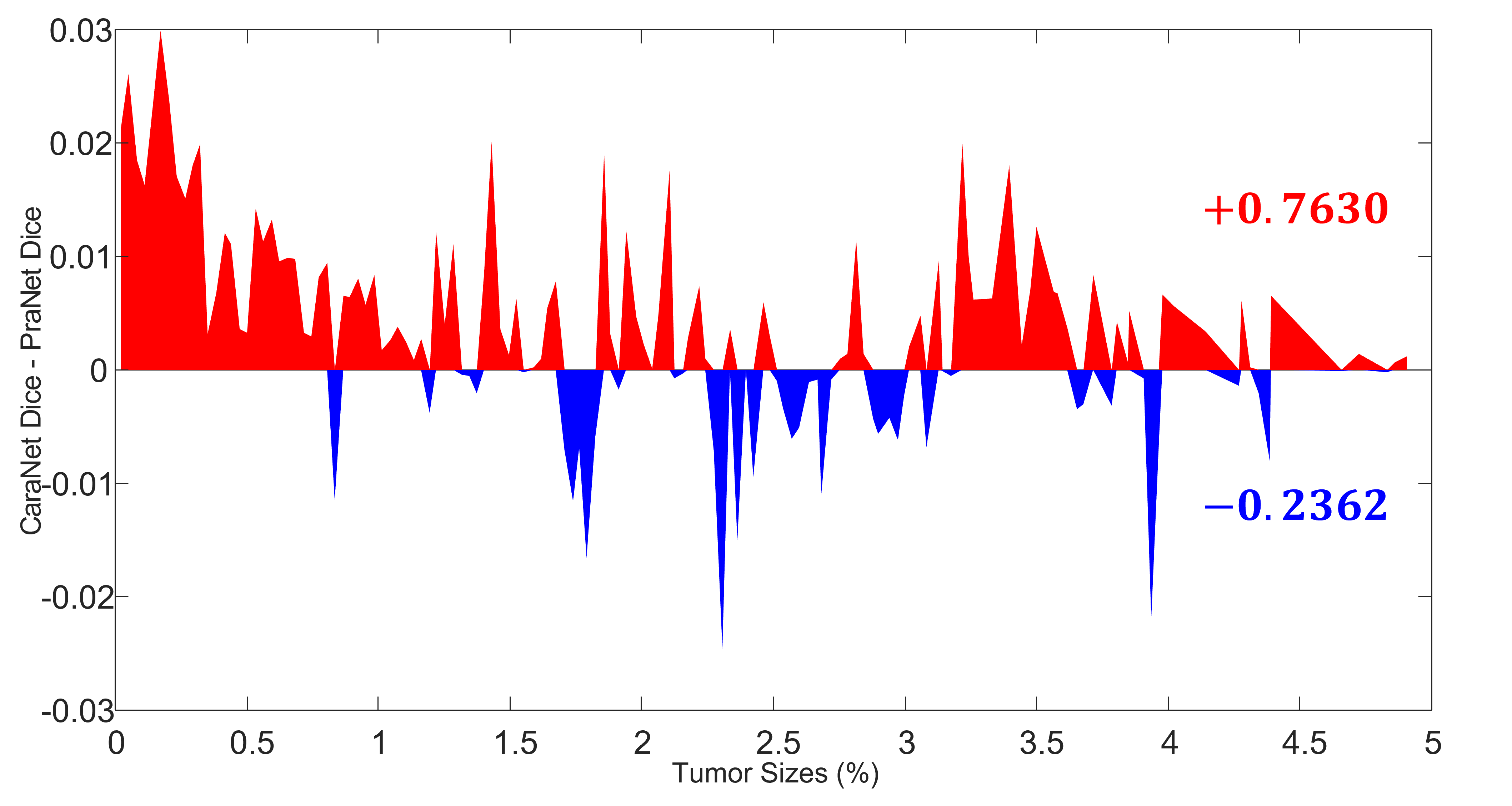
The difference between results of CaraNet and PraNet:
Citation
@article{lou2021caranet,
title={CaraNet: Context Axial Reverse Attention Network for Segmentation of Small Medical Objects},
author={Ange Lou and Shuyue Guan and Murray Loew},
journal={arXiv preprint arXiv:2108.07368},
year={2021}
}
@article{lou2021cfpnet,
title={CFPNet: Channel-wise Feature Pyramid for Real-Time Semantic Segmentation},
author={Lou, Ange and Loew, Murray},
journal={arXiv preprint arXiv:2103.12212},
year={2021}
}
GitHub
https://github.com/AngeLouCN/CaraNethttps://github.com/AngeLouCN/CaraNet




