miceforest
Fast, memory efficient Multiple Imputation by Chained Equations (MICE) with random forests. It can impute categorical and numeric data without much setup, and has an array of diagnostic plots available.
Package Meta
miceforest has 4 main classes which the user will interact with:
KernelDataSet- a kernel data set is a dataset on which the mice
algorithm is performed. Models are saved inside the instance, which
can also be called on to impute new data. Several plotting methods
are included to run diagnostics on the imputed data.MultipleImputedKernel- a collection ofKernelDataSets. Has
additional methods for accessing and comparing multiple kernel
datasets together.ImputedDataSet- a single dataset that has been imputed. These are
returned afterimpute_new_data()is called.MultipleImputedDataSet- A collection of datasets that have been
imputed. Has additional methods for comparing the imputations
between datasets.
This package can be installed using either pip or conda, through
conda-forge:
# Using pip
$ pip install miceforest
# Using conda
$ conda install -c conda-forge miceforest
You can also download the latest development version from this
repository. If you want to install from github with conda, you must
first run conda install pip git.
$ pip install git+https://github.com/AnotherSamWilson/miceforest.git
Using miceforest
We will be looking at a few simple examples of imputation. We need to
load the packages, and define the data:
import miceforest as mf
from sklearn.datasets import load_iris
import pandas as pd
import numpy as np
# Load data and introduce missing values
iris = pd.concat(load_iris(as_frame=True,return_X_y=True),axis=1)
iris['target'] = iris['target'].astype('category')
iris_amp = mf.ampute_data(iris,perc=0.25,random_state=1991)
Imputing a Single Dataset
If you only want to create a single imputed dataset, you can use
KernelDataSet:
# Create kernel.
kds = mf.KernelDataSet(
iris_amp,
save_all_iterations=True,
random_state=1991
)
# Run the MICE algorithm for 3 iterations
kds.mice(3)
# Return the completed kernel data
completed_data = kds.complete_data()
There are also an array of plotting functions available, these are
discussed below in the section Diagnostic
Plotting.
The plotting behavior between single imputed datasets and multi-imputed
datasets is slightly different.
Simple Example of Multiple Imputation
We can also create a class which contains multiple KernelDataSets,
along with easy ways to compare them:
# Create kernel.
kernel = mf.MultipleImputedKernel(
iris_amp,
datasets=4,
save_all_iterations=True,
random_state=1991
)
# Run the MICE algorithm for 3 iterations on each of the datasets
kernel.mice(3)
Printing the MultipleImputedKernel object will tell you some high
level information:
print(kernel)
## Class: MultipleImputedKernel
## Models Saved: Last Iteration
## Datasets: 4
## Iterations: 3
## Imputed Variables: 5
## save_all_iterations: True
Controlling Tree Growth
A very nice thing about random forests is that they are trivially
parallelizable. We can save a lot of time by setting the n_jobs
parameter in both the fit and predict methods for the random forests:
# Run the MICE algorithm for 2 more iterations on the kernel
kernel.mice(2,n_jobs=2)
Any other arguments may be passed to either class
(RandomForestClassifier,RandomForestRegressor). In our example, we
may not have saved much (if any) time. This is because there is overhead
with using multiple cores, and our data is very small.
Creating a Custom Imputation Schema
It is possible to customize our imputation procedure by variable. By
passing a named list to variable_schema, you can specify the
predictors for each variable to impute. You can also select which
variables should be imputed using mean matching, as well as the mean
matching candidates, by passing a dict tomean_match_candidates:
var_sch = {
'sepal width (cm)': ['target','petal width (cm)'],
'petal width (cm)': ['target','sepal length (cm)']
}
var_mmc = {
'sepal width (cm)': 5,
'petal width (cm)': 0
}
cust_kernel = mf.MultipleImputedKernel(
iris_amp,
datasets=3,
variable_schema=var_sch,
mean_match_candidates=var_mmc
)
cust_kernel.mice(2)
Imputing New Data with Existing Models
Multiple Imputation can take a long time. If you wish to impute a
dataset using the MICE algorithm, but don’t have time to train new
models, it is possible to impute new datasets using a
MultipleImputedKernel object. The impute_new_data() function uses
the random forests collected by MultipleImputedKernel to perform
multiple imputation without updating the random forest at each
iteration:
# Our 'new data' is just the first 15 rows of iris_amp
new_data = iris_amp.iloc[range(15)]
new_data_imputed = kernel.impute_new_data(new_data=new_data)
print(new_data_imputed)
## Class: MultipleImputedDataSet
## Datasets: 4
## Iterations: 5
## Imputed Variables: 5
## save_all_iterations: False
All of the imputation parameters (variable_schema,
mean_match_candidates, etc) will be carried over from the original
MultipleImputedKernel object. When mean matching, the candidate values
are pulled from the original kernel dataset. To impute new data, the
save_models parameter in MultipleImputedKernel must be > 0. If
save_models == 1, the model from the latest iteration is saved for
each variable. If save_models > 1, the model from each iteration is
saved. This allows for new data to be imputed in a more similar fashion
to the original mice procedure.
Diagnostic Plotting
As of now, miceforest has four diagnostic plots available.
Distribution of Imputed-Values
We probably want to know how the imputed values are distributed. We can
plot the original distribution beside the imputed distributions in each
dataset by using the plot_imputed_distributions method of an
MultipleImputedKernel object:
kernel.plot_imputed_distributions(wspace=0.3,hspace=0.3)
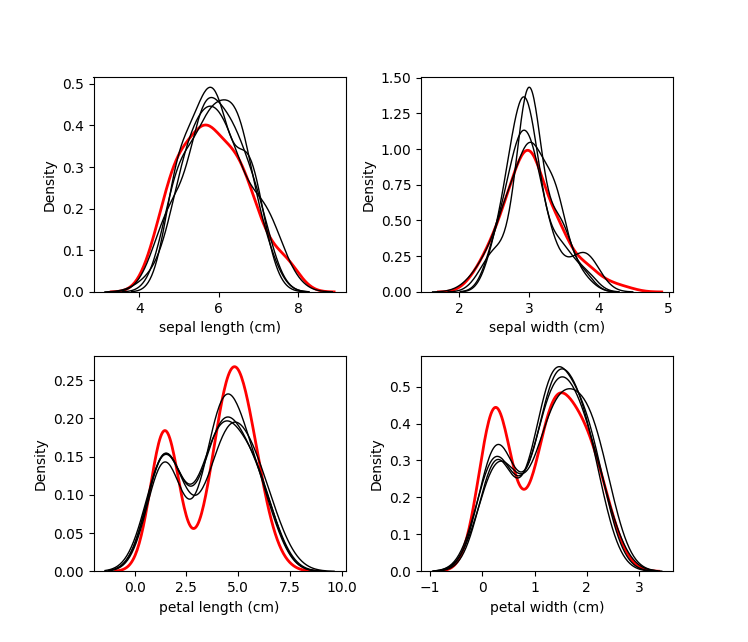
The red line is the original data, and each black line are the imputed
values of each dataset.
Convergence of Correlation
We are probably interested in knowing how our values between datasets
converged over the iterations. The plot_correlations method shows you
a boxplot of the correlations between imputed values in every
combination of datasets, at each iteration. This allows you to see how
correlated the imputations are between datasets, as well as the
convergence over iterations:
kernel.plot_correlations()

Variable Importance
We also may be interested in which variables were used to impute each
variable. We can plot this information by using the
plot_feature_importance method.
kernel.plot_feature_importance(annot=True,cmap="YlGnBu",vmin=0, vmax=1)
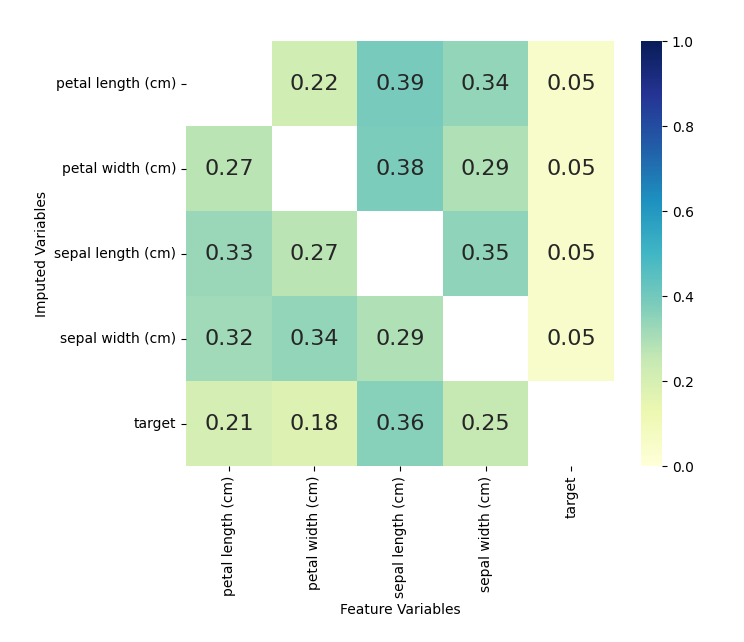
The numbers shown are returned from the sklearn random forest
_feature_importance attribute. Each square represents the importance
of the column variable in imputing the row variable.
Mean Convergence
If our data is not missing completely at random, we may see that it
takes a few iterations for our models to get the distribution of
imputations right. We can plot the average value of our imputations to
see if this is occurring:
kernel.plot_mean_convergence(wspace=0.3, hspace=0.4)
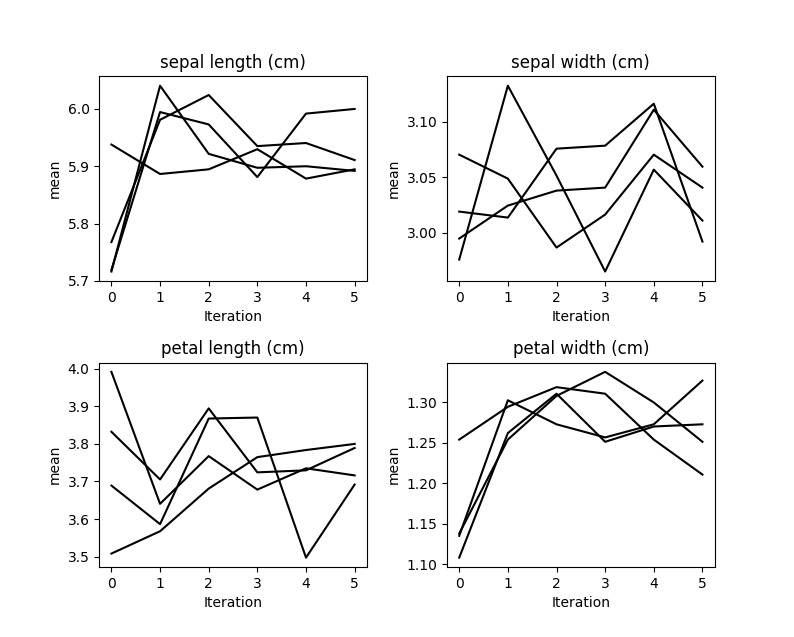
Our data was missing completely at random, so we don’t see any
convergence occurring here.
Using the Imputed Data
To return the imputed data simply use the complete_data method:
dataset_1 = kernel.complete_data(0)
This will return a single specified dataset. Multiple datasets are
typically created so that some measure of confidence around each
prediction can be created.
Since we know what the original data looked like, we can cheat and see
how well the imputations compare to the original data:
acclist = []
for iteration in range(kernel.iteration_count()+1):
target_na_count = kernel.na_counts['target']
compdat = kernel.complete_data(dataset=0,iteration=iteration)
# Record the accuract of the imputations of target.
acclist.append(
round(1-sum(compdat['target'] != iris['target'])/target_na_count,2)
)
# acclist shows the accuracy of the imputations
# over the iterations.
print(acclist)
## [0.32, 0.76, 0.78, 0.81, 0.86, 0.86]
In this instance, we went from a ~32% accuracy (which is expected with
random sampling) to an accuracy of ~86%. We managed to replace the
missing target values with a pretty high degree of accuracy!
The MICE Algorithm
Multiple Imputation by Chained Equations ‘fills in’ (imputes) missing
data in a dataset through an iterative series of predictive models. In
each iteration, each specified variable in the dataset is imputed using
the other variables in the dataset. These iterations should be run until
it appears that convergence has been met.
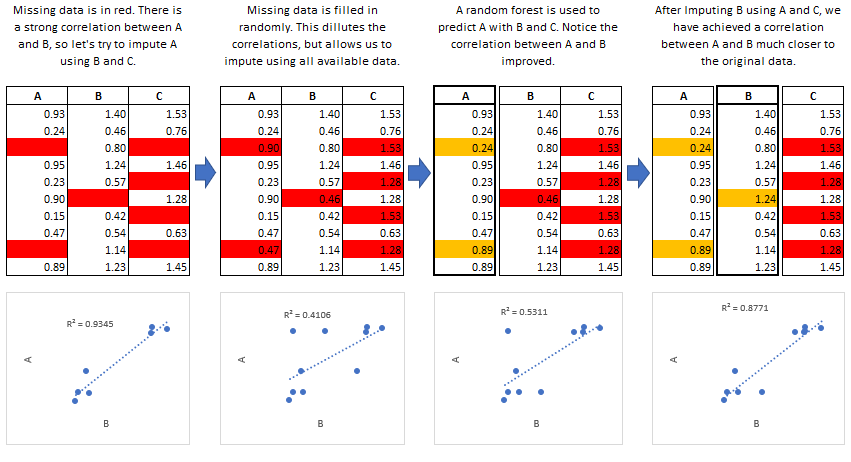
This process is continued until all specified variables have been
imputed. Additional iterations can be run if it appears that the average
imputed values have not converged, although no more than 5 iterations
are usually necessary.
Common Use Cases
Data Leakage:
MICE is particularly useful if missing values are associated with the
target variable in a way that introduces leakage. For instance, let’s
say you wanted to model customer retention at the time of sign up. A
certain variable is collected at sign up or 1 month after sign up. The
absence of that variable is a data leak, since it tells you that the
customer did not retain for 1 month.
Funnel Analysis:
Information is often collected at different stages of a ‘funnel’. MICE
can be used to make educated guesses about the characteristics of
entities at different points in a funnel.
Confidence Intervals:
MICE can be used to impute missing values, however it is important to
keep in mind that these imputed values are a prediction. Creating
multiple datasets with different imputed values allows you to do two
types of inference:
- Imputed Value Distribution: A profile can be built for each imputed
value, allowing you to make statements about the likely distribution
of that value. - Model Prediction Distribution: With multiple datasets, you can build
multiple models and create a distribution of predictions for each
sample. Those samples with imputed values which were not able to be
imputed with much confidence would have a larger variance in their
predictions.
Predictive Mean Matching
miceforest can make use of a procedure called predictive mean matching
(PMM) to select which values are imputed. PMM involves selecting a
datapoint from the original, nonmissing data which has a predicted value
close to the predicted value of the missing sample. The closest N
(mean_match_candidates parameter) values are chosen as candidates,
from which a value is chosen at random. This can be specified on a
column-by-column basis. Going into more detail from our example above,
we see how this works in practice:

This method is very useful if you have a variable which needs imputing
which has any of the following characteristics:
- Multimodal
- Integer
- Skewed
Effects of Mean Matching
As an example, let’s construct a dataset with some of the above
characteristics:
randst = np.random.RandomState(1991)
# random uniform variable
nrws = 1000
uniform_vec = randst.uniform(size=nrws)
def make_bimodal(mean1,mean2,size):
bimodal_1 = randst.normal(size=nrws, loc=mean1)
bimodal_2 = randst.normal(size=nrws, loc=mean2)
bimdvec = []
for i in range(size):
bimdvec.append(randst.choice([bimodal_1[i], bimodal_2[i]]))
return np.array(bimdvec)
# Make 2 Bimodal Variables
close_bimodal_vec = make_bimodal(2,-2,nrws)
far_bimodal_vec = make_bimodal(3,-3,nrws)
# Highly skewed variable correlated with Uniform_Variable
skewed_vec = np.exp(uniform_vec*randst.uniform(size=nrws)*3) + randst.uniform(size=nrws)*3
# Integer variable correlated with Close_Bimodal_Variable and Uniform_Variable
integer_vec = np.round(uniform_vec + close_bimodal_vec/3 + randst.uniform(size=nrws)*2)
# Make a DataFrame
dat = pd.DataFrame(
{
'uniform_var':uniform_vec,
'close_bimodal_var':close_bimodal_vec,
'far_bimodal_var':far_bimodal_vec,
'skewed_var':skewed_vec,
'integer_var':integer_vec
}
)
# Ampute the data.
ampdat = mf.ampute_data(dat,perc=0.25,random_state=randst)
# Plot the original data
import seaborn as sns
import matplotlib.pyplot as plt
g = sns.PairGrid(dat)
g.map(plt.scatter,s=5)
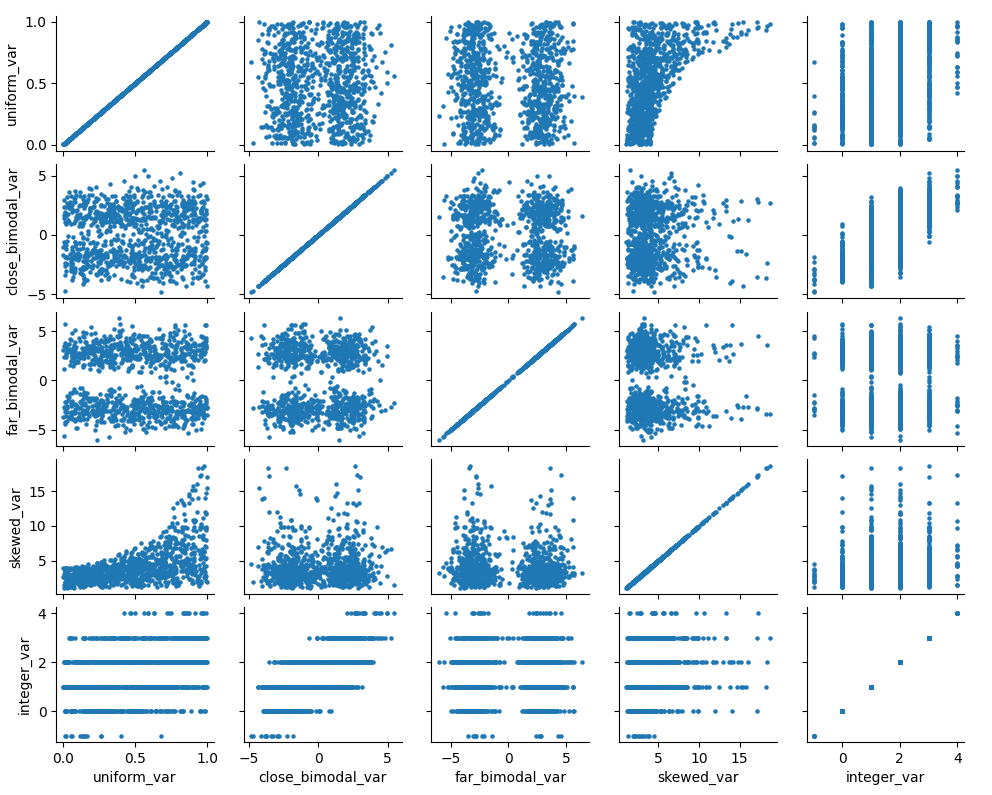 We can see how our variables are distributed and correlated in the graph
above. Now let’s run our imputation process twice, once using mean
matching, and once using the model prediction.
We can see how our variables are distributed and correlated in the graph
above. Now let’s run our imputation process twice, once using mean
matching, and once using the model prediction.
kernelmeanmatch <- mf.MultipleImputedKernel(ampdat,mean_match_candidates=5)
kernelmodeloutput <- mf.MultipleImputedKernel(ampdat,mean_match_candidates=0)
kernelmeanmatch.mice(5)
kernelmodeloutput.mice(5)
Let’s look at the effect on the different variables.
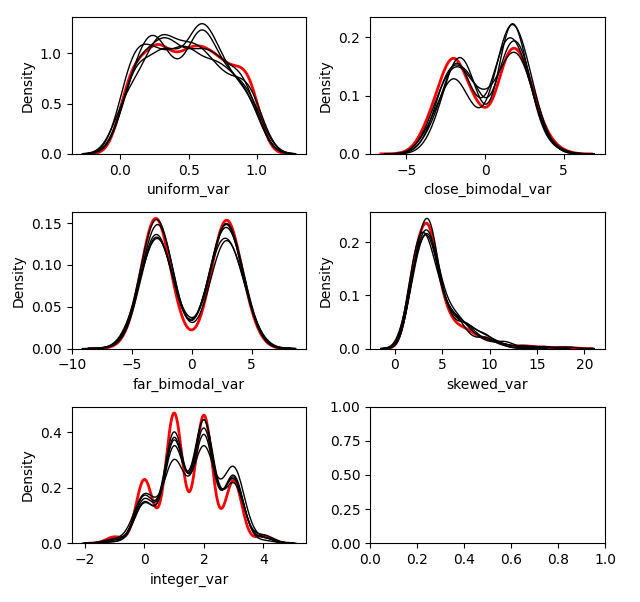
With Mean Matching
kernelmeanmatch.plot_imputed_distributions(wspace=0.2,hspace=0.4)
Without Mean Matching
kernelmodeloutput.plot_imputed_distributions(wspace=0.2,hspace=0.4)
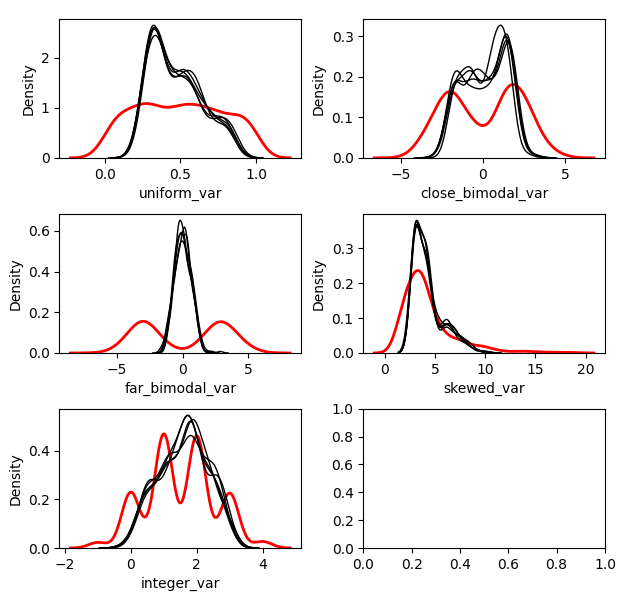
You can see the effects that mean matching has, depending on the
distribution of the data. Simply returning the value from the model
prediction, while it may provide a better ‘fit’, will not provide
imputations with a similair distribution to the original. This may be
beneficial, depending on your goal.








