AlphaFold
This package provides an implementation of the inference pipeline of AlphaFold v2.0. This is a completely new model that was entered in CASP14 and published in Nature. For simplicity, we refer to this model as AlphaFold throughout the rest of this document.
Any publication that discloses findings arising from using this source code or the model parameters should cite the AlphaFold paper.
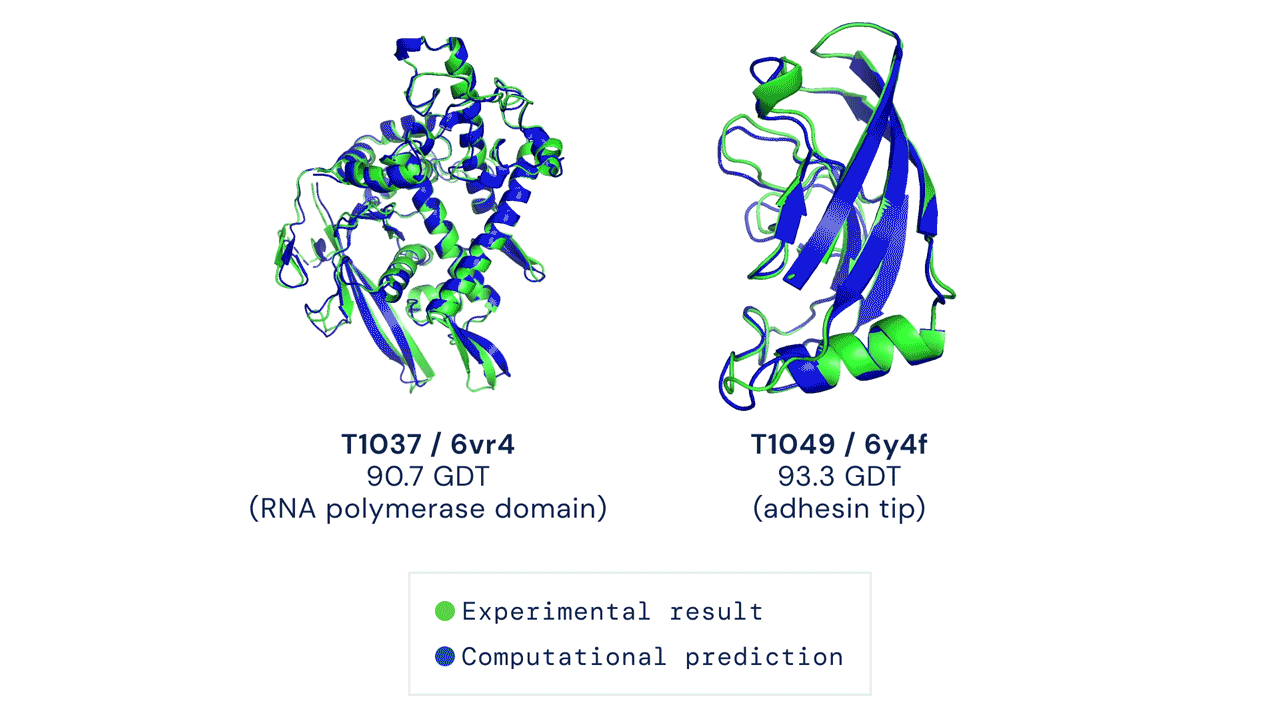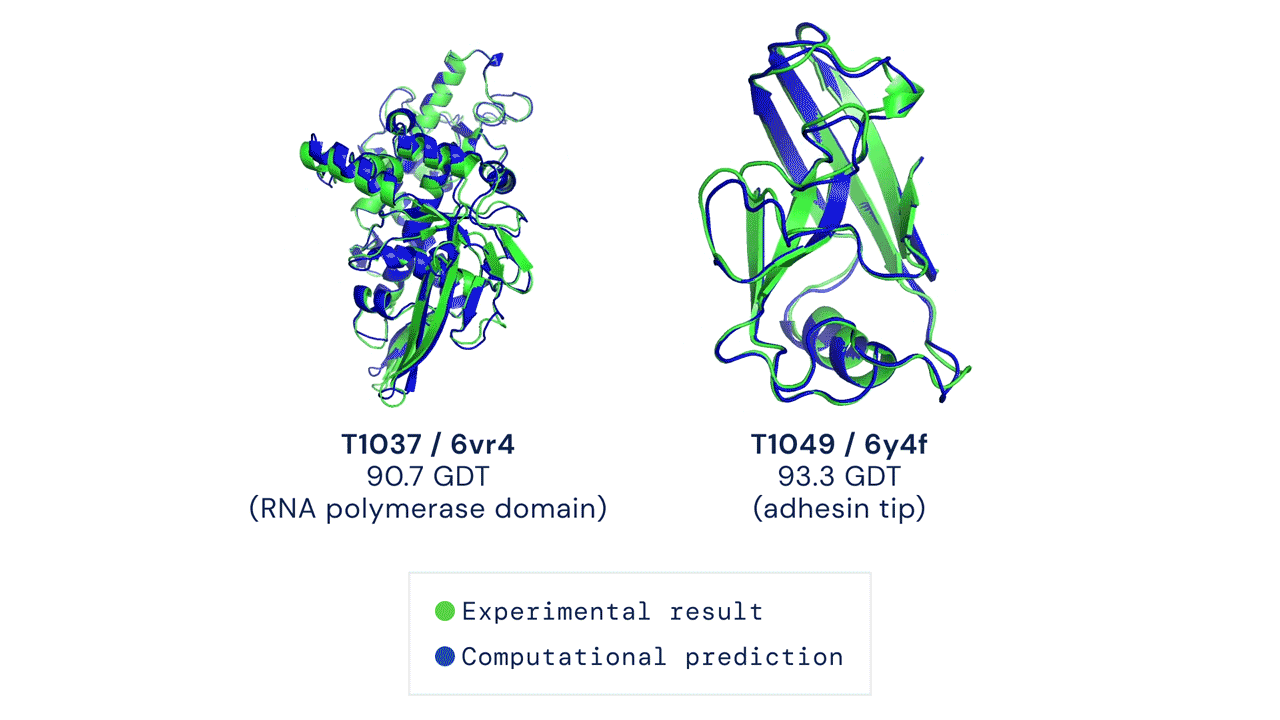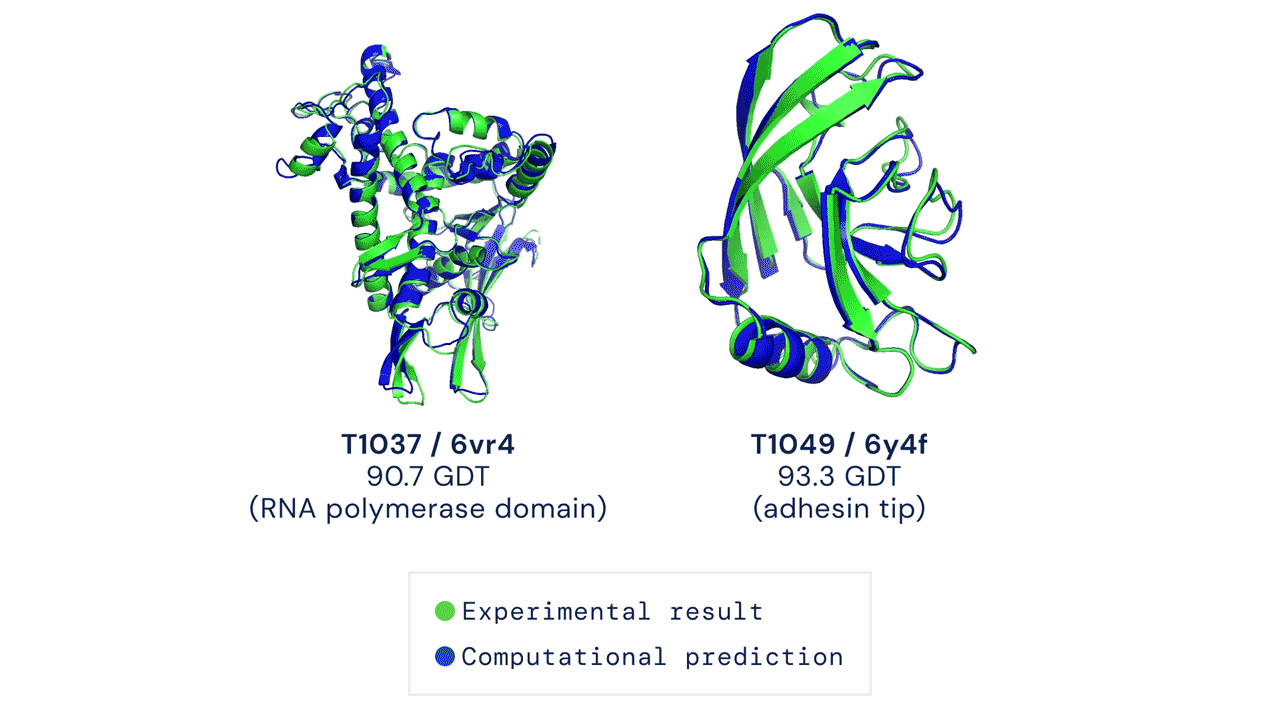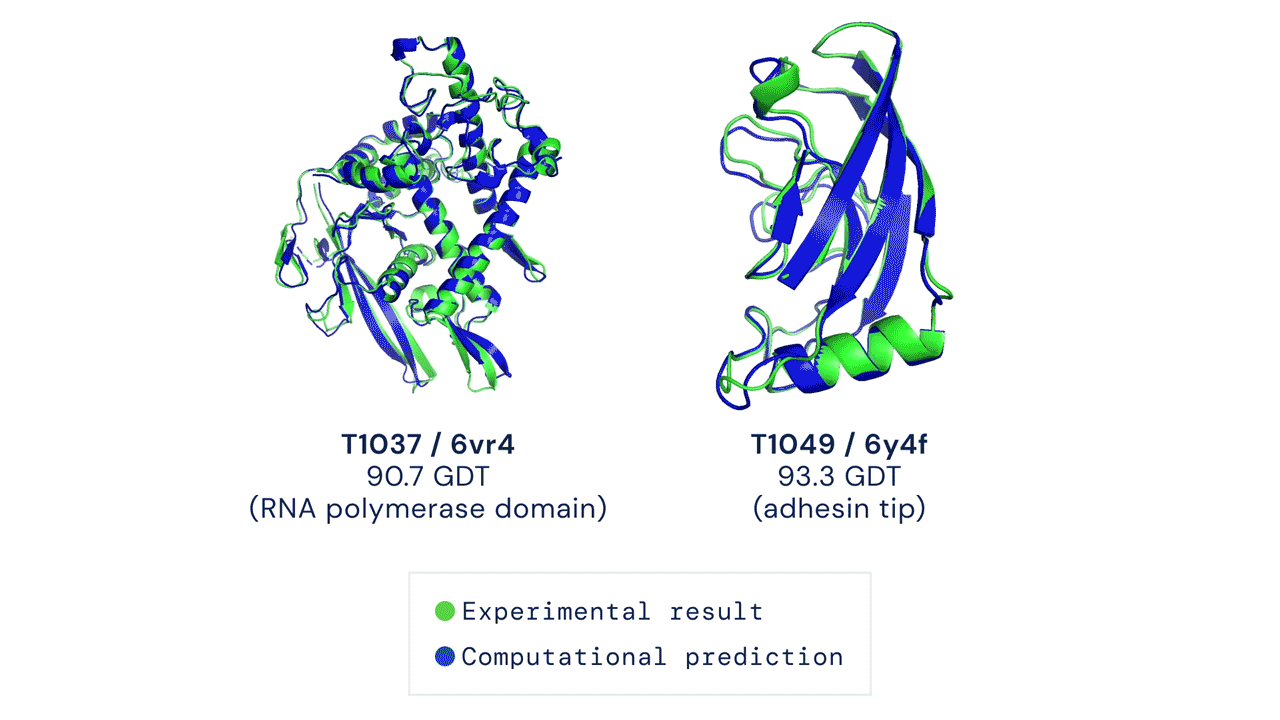
First time setup
The following steps are required in order to run AlphaFold:
-
Install Docker.
- Install
NVIDIA Container Toolkit
for GPU support. - Setup running
Docker as a non-root user.
- Install
-
Download genetic databases (see below).
-
Download model parameters (see below).
-
Check that AlphaFold will be able to use a GPU by running:
docker run --rm --gpus all nvidia/cuda:11.0-base nvidia-smiThe output of this command should show a list of your GPUs. If it doesn't,
check if you followed all steps correctly when setting up the
NVIDIA Container Toolkit
or take a look at the following
NVIDIA Docker issue.
Genetic databases
This step requires rsync and aria2c to be installed on your machine.
AlphaFold needs multiple genetic (sequence) databases to run:
We provide a script scripts/download_all_data.sh that can be used to download
and set up all of these databases. This should take 8–12 hours.
:ledger: Note: The total download size is around 428 GB and the total size
when unzipped is 2.2 TB. Please make sure you have a large enough hard drive
space, bandwidth and time to download.
This script will also download the model parameter files. Once the script has
finished, you should have the following directory structure:
$DOWNLOAD_DIR/ # Total: ~ 2.2 TB (download: 428 GB)
bfd/ # ~ 1.8 TB (download: 271.6 GB)
# 6 files.
mgnify/ # ~ 64 GB (download: 32.9 GB)
mgy_clusters.fa
params/ # ~ 3.5 GB (download: 3.5 GB)
# 5 CASP14 models,
# 5 pTM models,
# LICENSE,
# = 11 files.
pdb70/ # ~ 56 GB (download: 19.5 GB)
# 9 files.
pdb_mmcif/ # ~ 206 GB (download: 46 GB)
mmcif_files/
# About 180,000 .cif files.
obsolete.dat
uniclust30/ # ~ 87 GB (download: 24.9 GB)
uniclust30_2018_08/
# 13 files.
uniref90/ # ~ 59 GB (download: 29.7 GB)
uniref90.fasta
Model parameters
While the AlphaFold code is licensed under the Apache 2.0 License, the AlphaFold
parameters are made available for non-commercial use only under the terms of the
CC BY-NC 4.0 license. Please see the Disclaimer below
for more detail.
The AlphaFold parameters are available from
https://storage.googleapis.com/alphafold/alphafold_params_2021-07-14.tar, and
are downloaded as part of the scripts/download_all_data.sh script. This script
will download parameters for:
- 5 models which were used during CASP14, and were extensively validated for
structure prediction quality (see Jumper et al. 2021, Suppl. Methods 1.12
for details). - 5 pTM models, which were fine-tuned to produce pTM (predicted TM-score) and
predicted aligned error values alongside their structure predictions (see
Jumper et al. 2021, Suppl. Methods 1.9.7 for details).
Running AlphaFold
The simplest way to run AlphaFold is using the provided Docker script. This
was tested on Google Cloud with a machine using the nvidia-gpu-cloud-image
with 12 vCPUs, 85 GB of RAM, a 100 GB boot disk, the databases on an additional
3 TB disk, and an A100 GPU.
-
Clone this repository and
cdinto it.git clone https://github.com/deepmind/alphafold.git -
Modify
DOWNLOAD_DIRindocker/run_docker.pyto be the path to the
directory containing the downloaded databases. -
Build the Docker image:
docker build -f docker/Dockerfile -t alphafold . -
Install the
run_docker.pydependencies. Note: You may optionally wish to
create a
Python Virtual Environment
to prevent conflicts with your system's Python environment.pip3 install -r docker/requirements.txt -
Run
run_docker.pypointing to a FASTA file containing the protein sequence
for which you wish to predict the structure. If you are predicting the
structure of a protein that is already in PDB and you wish to avoid using it
as a template, thenmax_template_datemust be set to be before the release
date of the structure. For example, for the T1050 CASP14 target:python3 docker/run_docker.py --fasta_paths=T1050.fasta --max_template_date=2020-05-14By default, Alphafold will attempt to use all visible GPU devices. To use a
subset, specify a comma-separated list of GPU UUID(s) or index(es) using the
--gpu_devicesflag. See
GPU enumeration
for more details. -
You can control AlphaFold speed / quality tradeoff by adding either
--preset=full_dbsor--preset=casp14to the run command. We provide the
following presets:- casp14: This preset uses the same settings as were used in CASP14.
It runs with all genetic databases and with 8 ensemblings. - full_dbs: The model in this preset is 8 times faster than the
casp14preset with a very minor quality drop (-0.1 average GDT drop on
CASP14 domains). It runs with all genetic databases and with no
ensembling.
Running the command above with the
casp14preset would look like this:python3 docker/run_docker.py --fasta_paths=T1050.fasta --max_template_date=2020-05-14 --preset=casp14 - casp14: This preset uses the same settings as were used in CASP14.
AlphaFold output
The outputs will be in a subfolder of output_dir in run_docker.py. They
include the computed MSAs, unrelaxed structures, relaxed structures, ranked
structures, raw model outputs, prediction metadata, and section timings. The
output_dir directory will have the following structure:
output_dir/
features.pkl
ranked_{0,1,2,3,4}.pdb
ranking_debug.json
relaxed_model_{1,2,3,4,5}.pdb
result_model_{1,2,3,4,5}.pkl
timings.json
unrelaxed_model_{1,2,3,4,5}.pdb
msas/
bfd_uniclust_hits.a3m
mgnify_hits.sto
uniref90_hits.sto
The contents of each output file are as follows:
features.pkl– Apicklefile containing the input feature Numpy arrays
used by the models to produce the structures.unrelaxed_model_*.pdb– A PDB format text file containing the predicted
structure, exactly as outputted by the model.relaxed_model_*.pdb– A PDB format text file containing the predicted
structure, after performing an Amber relaxation procedure on the unrelaxed
structure prediction, see Jumper et al. 2021, Suppl. Methods 1.8.6 for
details.ranked_*.pdb– A PDB format text file containing the relaxed predicted
structures, after reordering by model confidence. Hereranked_0.pdbshould
contain the prediction with the highest confidence, andranked_4.pdbthe
prediction with the lowest confidence. To rank model confidence, we use
predicted LDDT (pLDDT), see Jumper et al. 2021, Suppl. Methods 1.9.6 for
details.ranking_debug.json– A JSON format text file containing the pLDDT values
used to perform the model ranking, and a mapping back to the original model
names.timings.json– A JSON format text file containing the times taken to run
each section of the AlphaFold pipeline.msas/- A directory containing the files describing the various genetic
tool hits that were used to construct the input MSA.result_model_*.pkl– Apicklefile containing a nested dictionary of the
various Numpy arrays directly produced by the model. In addition to the
output of the structure module, this includes auxiliary outputs such as
distograms and pLDDT scores. If using the pTM models then the pTM logits
will also be contained in this file.
This code has been tested to match mean top-1 accuracy on a CASP14 test set with
pLDDT ranking over 5 model predictions (some CASP targets were run with earlier
versions of AlphaFold and some had manual interventions; see our forthcoming
publication for details). Some targets such as T1064 may also have high
individual run variance over random seeds.
Inferencing many proteins
The provided inference script is optimized for predicting the structure of a
single protein, and it will compile the neural network to be specialized to
exactly the size of the sequence, MSA, and templates. For large proteins, the
compile time is a negligible fraction of the runtime, but it may become more
significant for small proteins or if the multi-sequence alignments are already
precomputed. In the bulk inference case, it may make sense to use our
make_fixed_size function to pad the inputs to a uniform size, thereby reducing
the number of compilations required.
We do not provide a bulk inference script, but it should be straightforward to
develop on top of the RunModel.predict method with a parallel system for
precomputing multi-sequence alignments. Alternatively, this script can be run
repeatedly with only moderate overhead.
Note on reproducibility
AlphaFold's output for a small number of proteins has high inter-run variance,
and may be affected by changes in the input data. The CASP14 target T1064 is a
notable example; the large number of SARS-CoV-2-related sequences recently
deposited changes its MSA significantly. This variability is somewhat mitigated
by the model selection process; running 5 models and taking the most confident.
To reproduce the results of our CASP14 system as closely as possible you must
use the same database versions we used in CASP. These may not match the default
versions downloaded by our scripts.
For genetics:
- UniRef90:
v2020_01 - MGnify:
v2018_12 - Uniclust30: v2018_08
- BFD: only version available
For templates:
- PDB: (downloaded 2020-05-14)
- PDB70: (downloaded 2020-05-13)
An alternative for templates is to use the latest PDB and PDB70, but pass the
flag --max_template_date=2020-05-14, which restricts templates only to
structures that were available at the start of CASP14.
Citing this work
If you use the code or data in this package, please cite:
@Article{AlphaFold2021,
author = {Jumper, John and Evans, Richard and Pritzel, Alexander and Green, Tim and Figurnov, Michael and Ronneberger, Olaf and Tunyasuvunakool, Kathryn and Bates, Russ and {\v{Z}}{\'\i}dek, Augustin and Potapenko, Anna and Bridgland, Alex and Meyer, Clemens and Kohl, Simon A A and Ballard, Andrew J and Cowie, Andrew and Romera-Paredes, Bernardino and Nikolov, Stanislav and Jain, Rishub and Adler, Jonas and Back, Trevor and Petersen, Stig and Reiman, David and Clancy, Ellen and Zielinski, Michal and Steinegger, Martin and Pacholska, Michalina and Berghammer, Tamas and Bodenstein, Sebastian and Silver, David and Vinyals, Oriol and Senior, Andrew W and Kavukcuoglu, Koray and Kohli, Pushmeet and Hassabis, Demis},
journal = {Nature},
title = {Highly accurate protein structure prediction with {AlphaFold}},
year = {2021},
doi = {10.1038/s41586-021-03819-2},
note = {(Accelerated article preview)},
}




