FMST-PT
Implementation of paper FMSF-PT: A fast 3D medical image segmentation framework guided by phased tasks using pytorch.
The reasoning speed of traditional networks such as swinnet, unetr, nnunet and nnformer is FPS < 1, but our reasoning speed is super fast. You can get any 3D medical impact data after analysis through FPS > 15 processing speed. Tested on FPS 2017_ consenuse>30, FPS_ attention>20, FPS_ edge>15.
And we propose a dice evaluation for compressed data mask, attndice and a corresponding loss function attnloss. When the mask data is 0 or 1, attndice = dice. When the value is between 0 and 1, attndice > dice, which can more accurately judge the edge accuracy of compressed mask.
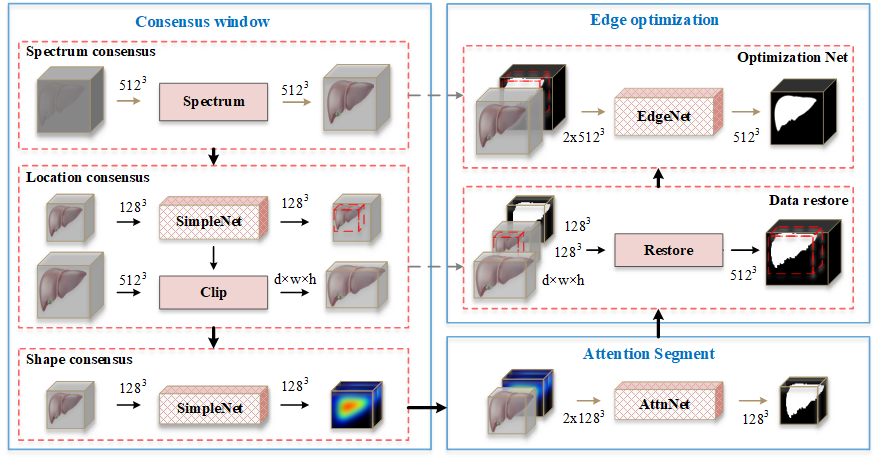
We disclosed some codes, including network structure in ./model/fmst.py, It contains three networks: “snet”, “attnett” and “edgenet”, which correspond to the three stages in the framework.
You can use our program to carry out secondary training one by one through the following commands(You need to modify the optimal weight position specified in the subsequent code after each training!):
python train_attention.py
python train_consensus.py
python train_edge.py
We uploaded the reasoning results on lits2017 for reference. Location: All stage, Consensus, Attention, Edge, and some of them are shown in the figure below:
sunet |
attnnet |
edgenet |
|---|---|---|
 |
 |
 |
We try to train and infer 14 classifications on btcv synapse data set based on the weight of lits 2017 liver 2 classification, which is only 2 hours on rtx3090, and give the output results of the visualization framework. We used 690 CT as the training set and 180 CT as the test set, and the reasoning speed of the model can still be maintained at FPS > 10.