impy is All You Need in Image Analysis
impy is an all-in-one image analysis library, equipped with parallel processing, GPU support, GUI based tools and so on.
The core array, ImgArray, is a subclass of numpy.ndarray, tagged with information such as
- image axes
- scale of each axis
- directory of the original image
- history of image processing
- and other image metadata
By making full use of them, impy provides super efficient tools of image analysis for you.
I'm also working on documentation for tutorials and API. Please take a look if you're interested in.
Code as fast as you speak
Almost all the functions, such as filtering, deconvolution, labeling, single molecule detection, and even those pure numpy functions, are aware of image metadata. They "know" which dimension corresponds to "z" axis, which axes they should iterate along or where to save the image. As a result, your code will be very concise:
import impy as ip
import numpy as np
img = ip.imread("path/to/image") # Read images with metadata.
img["z=3;t=0"].imshow() # Plot image slice at z=3 and t=0.
img_fil = img.gaussian_filter(sigma=2) # Paralell batch denoising. No more for loop!
img_prj = np.max(img_fil, axis="z") # Z-projection (numpy is aware of image axes!).
img_prj.imsave(f"Max-{img.name}") # Save in the same place. Don't spend time on searching for the directory!
Seamless interface between napari
napari is an interactive viewer for multi-dimensional images. impy has a simple and efficient interface with it, via the object ip.gui. Since ImgArray is tagged with image metadata, you don't have to care about axes or scales. Just run
ip.gui.add(img)
impy's viewer also provides many useful widgets and functions such as
- Excel-like table
- Compact file explorer
- interactive
matplotlibfigure canvas - cropping, duplication, measurement, filtering tools
Extend your function for batch processing
Already have a function for numpy? Decorate it with @ip.bind
@ip.bind
def imfilter(img, param=None):
# Your function here.
# Do something on a 2D or 3D image and return image, scalar or labels
return out
and it's ready for batch processing!
img.imfilter(param=1.0)
Making plugin is easy
Image analysis usually relies on manual handling, which has been discoraging people from programatic analysis on their data. But now, with @ip.gui.bind decorator, make plugin by yourself!
import matplotlib.pyplot as plt
from skimage.measure import moments
@ip.gui.bind
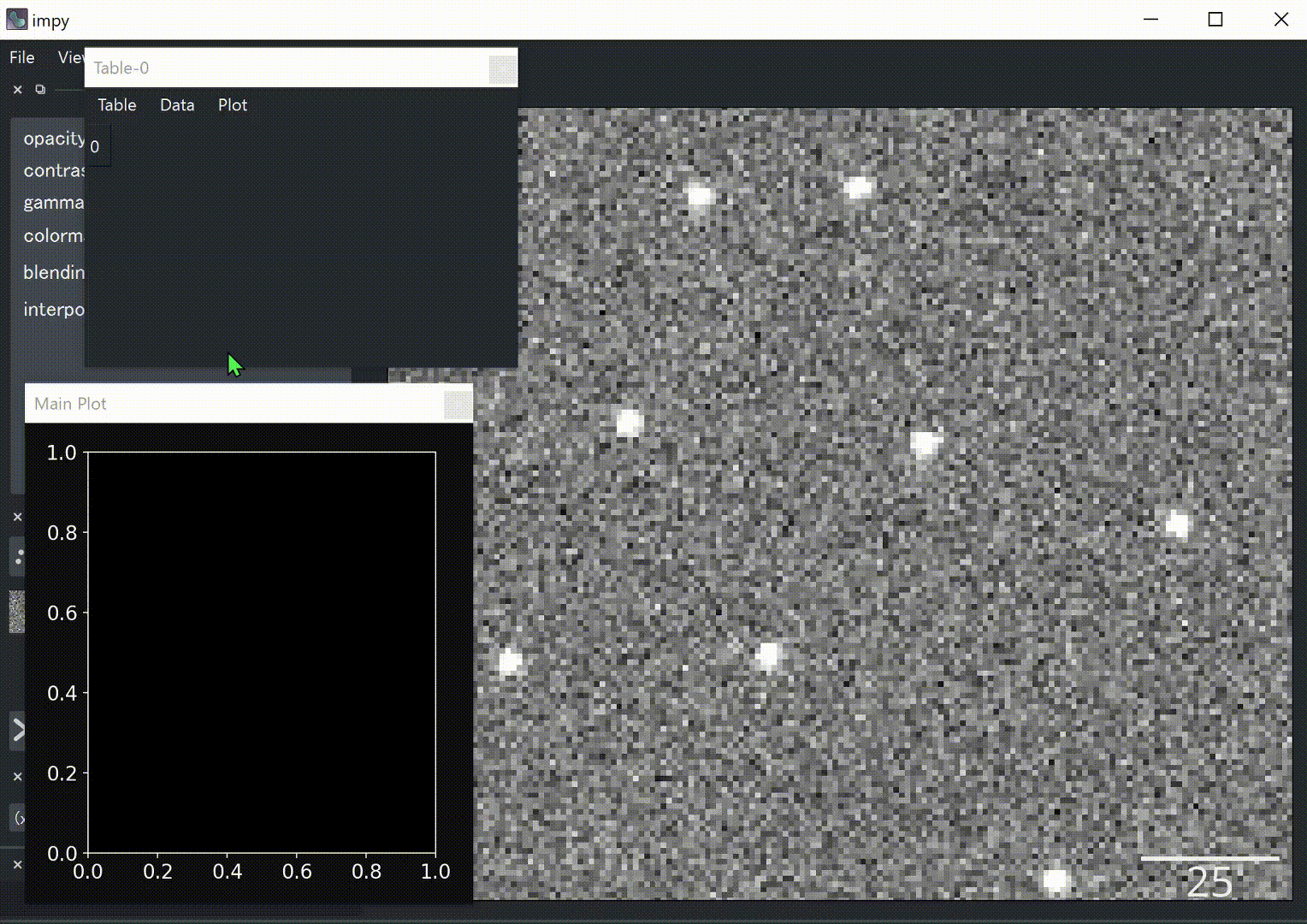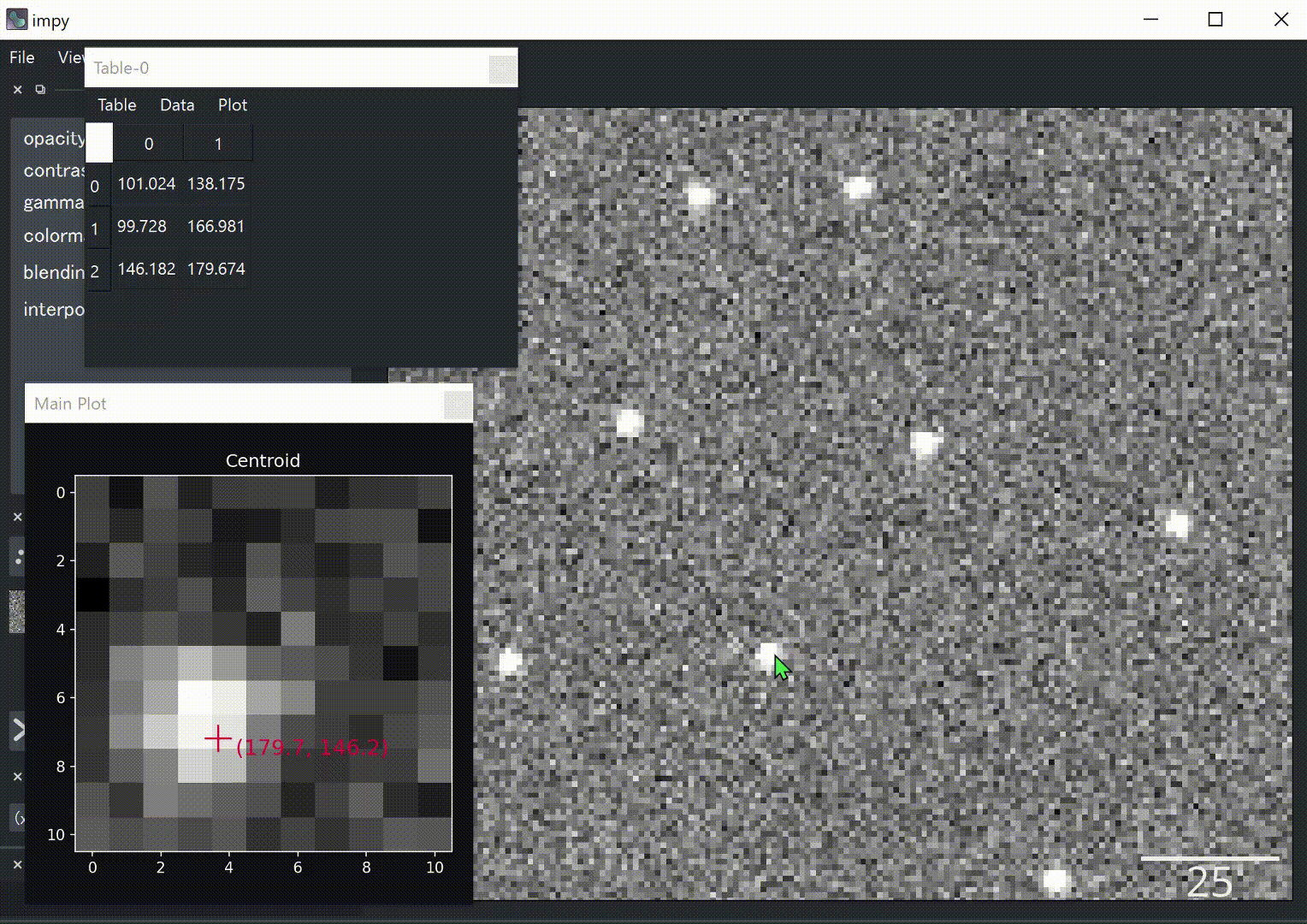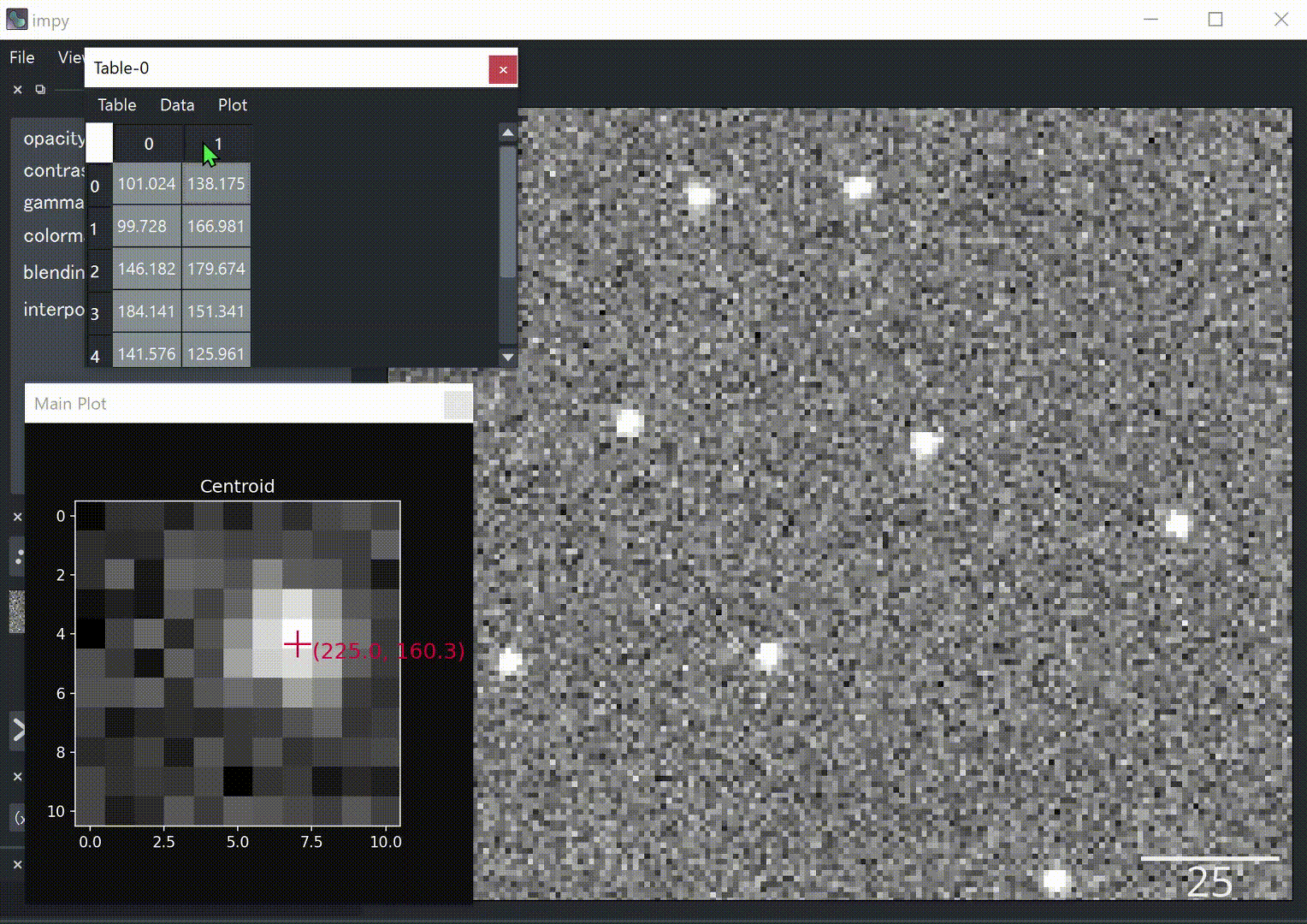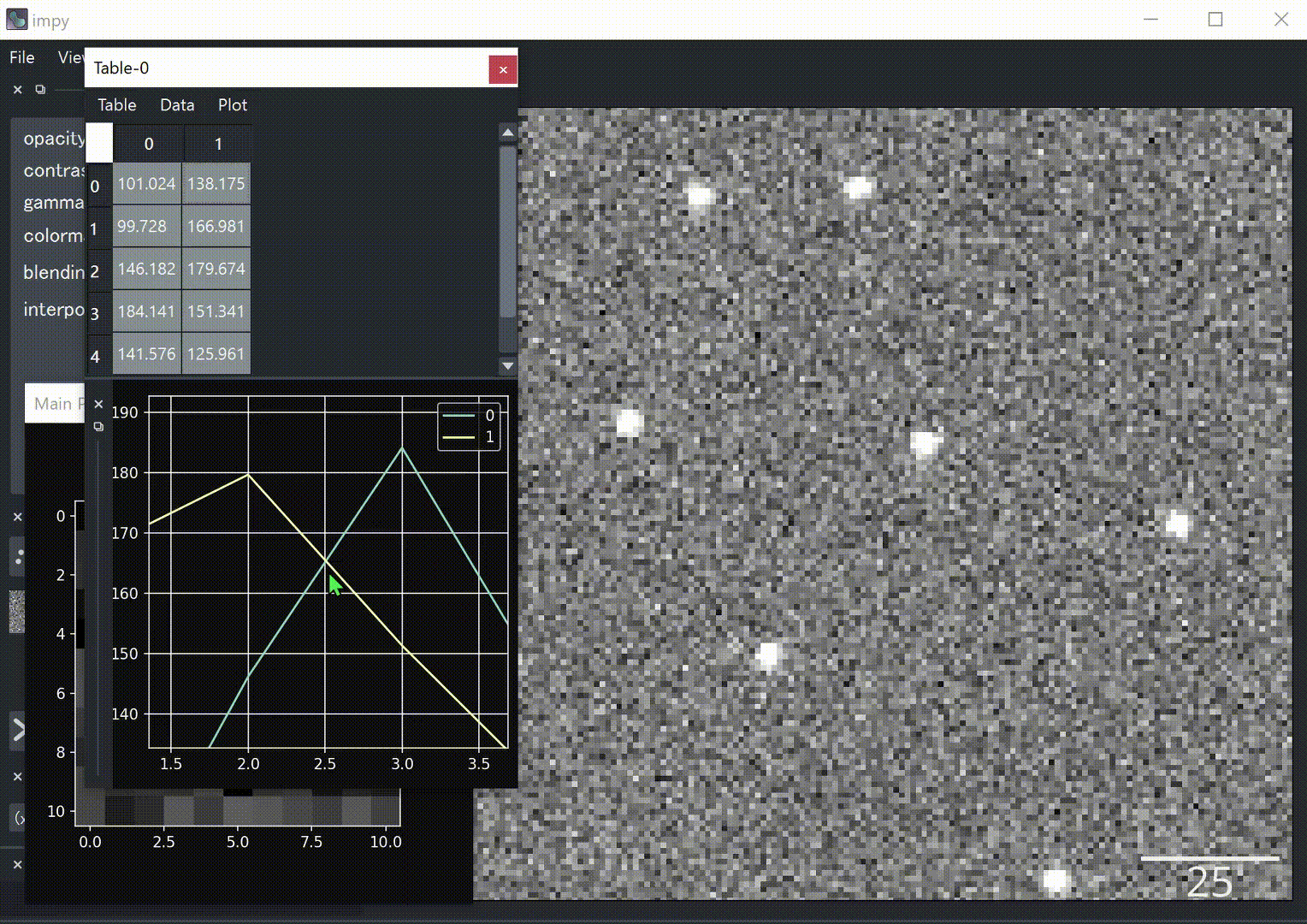
def func(gui): # the first argument will be ip.gui
img = gui.get("image") # Get the front image
y, x = gui.viewer.cursor.position # Get cursor position
y0 = int(y-5)
x0 = int(x-5)
img0 = img[y0:y0+11, x0:x0+11] # Get 11x11 region around cursor
# Calculate centroid.
M = moments(img0.value)
cy, cx = M[1, 0]/M[0, 0], M[0, 1]/M[0, 0]
# Plot centroid.
plt.figure()
plt.imshow(img0, cmap="gray")
plt.scatter([cx], [cy], s=360, color="crimson", marker="+")
plt.text(cx+0.5, cy+0.5, f"({cx+x0:.1f}, {cy+y0:.1f})", size="x-large", color="crimson")
plt.title("Centroid")
plt.show()
# Append centroid to table widget.
gui.table.append([cy+y0, cx+x0])
return
Installation
pip install git+https://github.com/hanjinliu/impy
or
git clone https://github.com/hanjinliu/impy
Depencencies
scikit-image>=0.18numpy>=1.17scipy>=1.6.3matplotlib>=3.3.4pandas>=1.3.1dask>=2021.6.0tifffile>=2021.6.14napari>=0.4.9
impy is partly dependent on numba, cupy, trackpy and dask-image. Please install these packages if needed.
:warning: Image processing algorithms in ImgArray are almost stable so that their behavior will not change a lot. However, since napari is under development and I'm groping for better UI right now, any functions that currently implemented in impy viewer may change or no longer work in the future. Make sure keeping napari and impy updated when you use.








